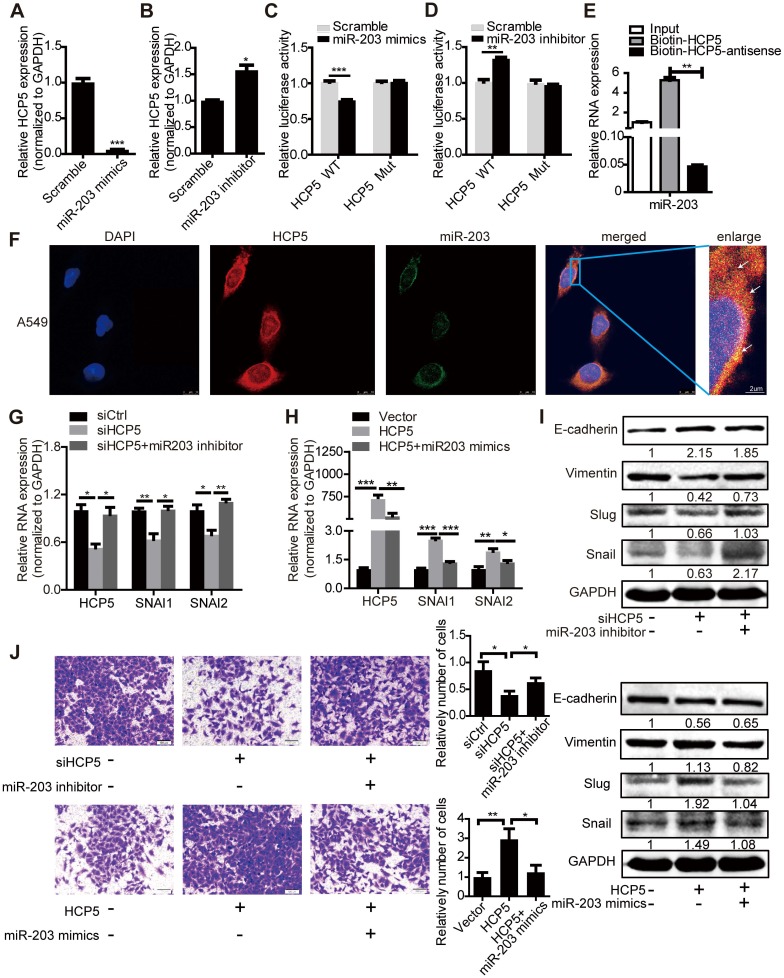
Figure 6.
HCP5 positively regulates EMT via the miR-203/SNAI axis. (A, B) The expression of HCP5 in A549 cells transfected with miR-203 mimics or inhibitors or negative control oligonucleotides. Data are shown as the mean ± S.E.M. of three independent experiments (two-tailed Student's t-test). (C, D) miR-203 mimics inhibit (C), whereas miR-203 inhibitors promote (D) Luciferase activity in A549 cells cotransfected with miR-203 mimics or inhibitors and luciferase reporters containing HCP5 or mutant transcript. Data are shown as the mean ± S.E.M. of three independent experiments (two-tailed Student's t-test). (E) MiR-203 was specifically pulled down by biotin-labelled HCP5 compared with HCP5-antisense in A549 cells. Data are shown as the mean ± S.E.M. of three independent experiments (two-tailed Student's t-test). (F) FISH assay HCP5 (red) is co-localized with miR-203 (green). The arrow indicates the approximate location. (G-I) The mRNA (G, H) or protein levels(I) of Slug, Snail, vimentin and E-cadherin when HCP5 knockdown A549 cells transfected with miR-203 inhibitors or HCP5-overexpressing A549 cells transfected with miR-203 mimics. Data are shown as the mean ± S.E.M. of three independent experiments (two-tailed Student's t-test). (J) Transwell assays to investigate the invasion ability of HCP5 knockdown A549 cells transfected with miR-203 inhibitors or HCP5-overexpressing A549 cells transfected with miR-203 mimics. Data are shown as the mean ± S.E.M. of three independent experiments (two-tailed Student's t-test). *P< 0.05, **P< 0.01, ***P < 0.001.