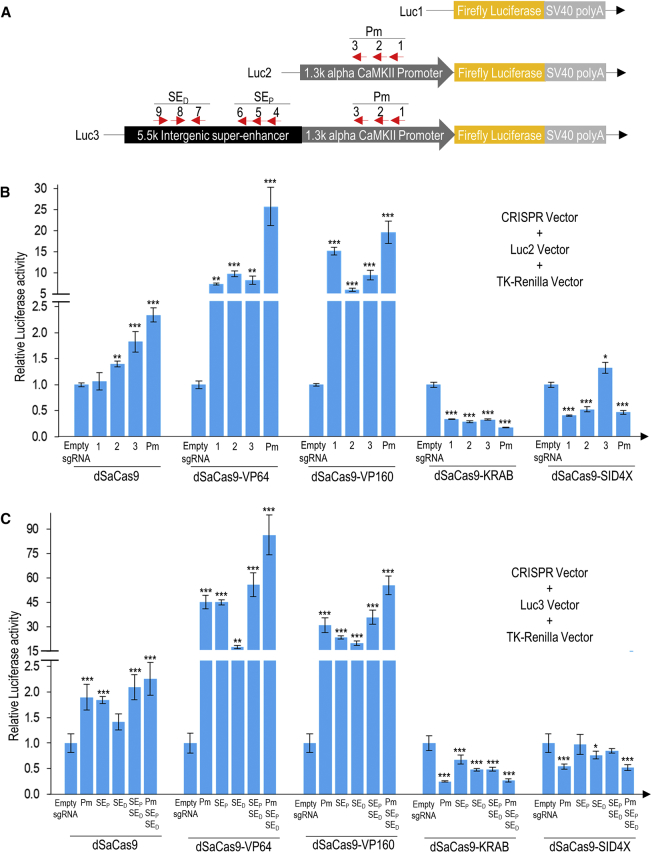
Figure 2.
Activation and Repression of Luciferase Activity in Mouse N2a Cells Mediated by Various Minimal CRISPRa and CRISPRi Transgenes
(A) Luciferase reporter vector construction. Luc1 is the promoterless pGL4.10[luc2] background control. Luc2 is a modified Luc1 vector that has been inserted with 1.3-kb mouse alpha CaMKII promoter amplified from the genomic DNA of N2a cells. Luc3 is a modified Luc1 vector that has been inserted with 6.8-kb mouse alpha CaMKII promoter and intergenic super-enhancer regions amplified from the genomic DNA of N2a cells. TK-Renilla was used as an internal normalization control in all luciferase reporter assay experiments. (B) Luciferase activity with dSaCas9-fused activators and repressors targeting mouse alpha CaMKII promoter. The CRISPRa or CRISPRi transgenes targeting the mouse alpha CaMKII promoter were co-transfected with Luc2 vector and TK-Renilla control vector. Three sgRNAs were designed to target three different binding sites on the promoter (Pm). Luciferase activities were measured when the CRISPRa or CRISPRi transgenes were used singly (site 1, 2, or 3) or in combination (sites 1, 2, and 3). (C) Luciferase activity with dSaCas9-fused activators and repressors targeting mouse alpha CaMKII promoter and intergenic super-enhancer. The sgRNAs targeting mouse alpha CaMKII promoter and intergenic super-enhancer were co-transfected with the Luc3 vector and TK-Renilla control vector. A total of nine sgRNAs was designed to target three different binding sites on the promoters (Pm), three on proximal (SEP) and three on distal intergenic super-enhancer sites (SED). All luciferase activities were normalized to the corresponding empty sgRNA. The statistical significance levels from control (empty sgRNA) are indicated as *p < 0.05, **p < 0.01, and ***p < 0.001. All data are presented as mean ± SD (n = 4).