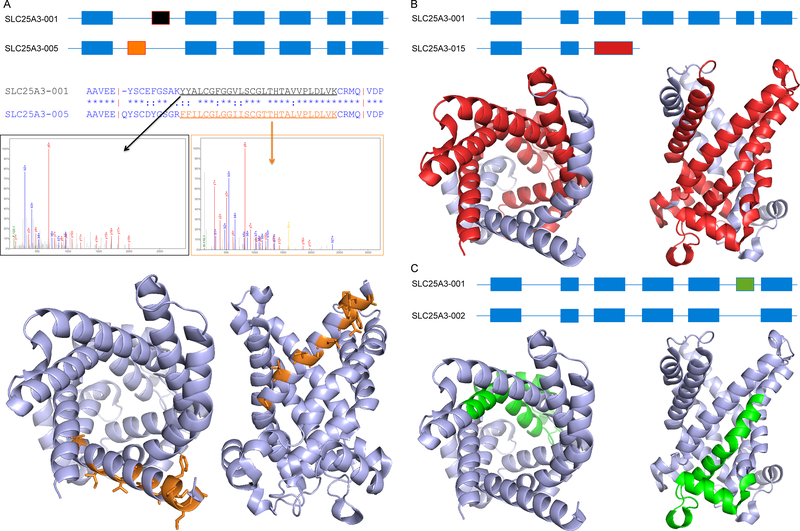
Figure 1. Types of alternative isoforms.
This figure presents three types of alternative variants defined using the gene SLC25A3, a mitochondrial phosphate carrier protein. In each case we show the effect at the transcript level and at the protein level. (A) Homologous exons. Above, schema of variant SCL25A3–005, which is generated from variant SCL25A3–001 via the substitution of exon 2a (black) by exon 2b (orange). The differing protein sequences are shown in the alignment below the transcript level comparison. Middle, example spectra for the two peptides that identify the two different alternative isoforms. Below, the likely effect on protein structure (shown in two views) for the similar gene SLC25A4 (PDB code: 1okc); residues that differ between the two isoforms are shown as orange sticks. The change to the structure and function is likely to be comparatively subtle: no residues are lost and most of the changes are found on the outside of the pore. (B) Non-homologous substitution. Above, schema of variant SCL25A3–015, which is generated from variant SCL25A3–001 via the substitution of exon 3 (the longer alternative exon is in red). Below; the likely effect on protein structure shown in two different views; residues that would be lost in the alternative isoforms are shown in red. (C) Indels. Above, schema of variant SCL25A3–002, which is generated from variant SCL25A3–001 via the skipping of exon 6 (green). Below, the likely structural effect of this loss of 28 amino acids is shown in two different views; residues that would be lost in the alternative isoforms are shown in green. The deletion would remove the base of the pore and parts of two different trans-membrane helices meaning that the trans-membrane sections would have to completely refold. Images generated with the PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.