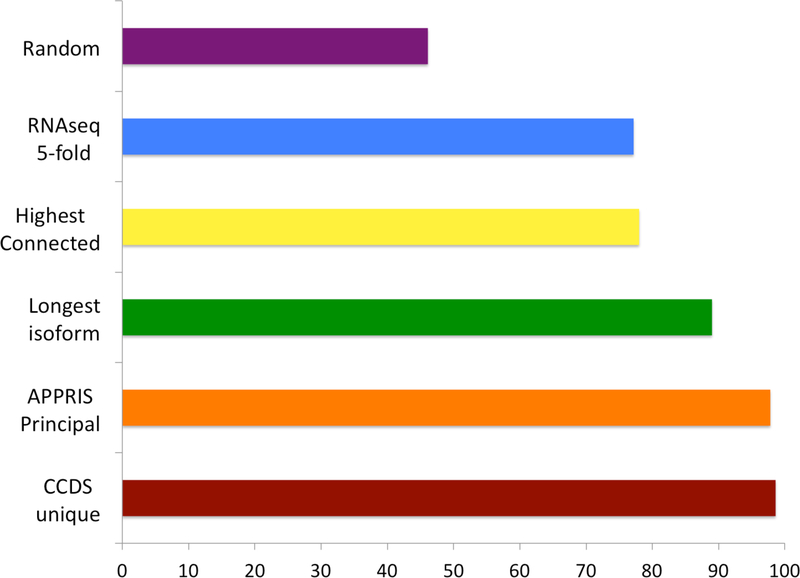
Figure 2. Coincidence between main proteomics isoforms and other reference isoforms.
The percentage of genes in which there was agreement between the reference isoform for a gene and the main proteomics isoform calculated from the proteomics experiments [36]. The comparison was made over all 5,011 genes from the same proteomics study for the longest isoform, over a subset of 3,331 genes with CCDS unique isoforms [41] for the CCDS comparison, over a subset of 4,186 genes with principal isoforms for the APPRIS comparison [43] and over a subset of 1,038 genes with five-fold dominant transcripts across all tissues for the RNAseq comparison [37]. The Highest Connected Isoform comparison was made using data from the paper that introduced the method [42]. A random selection of isoforms would have agreed with the main proteomics isoform 46% of the time.