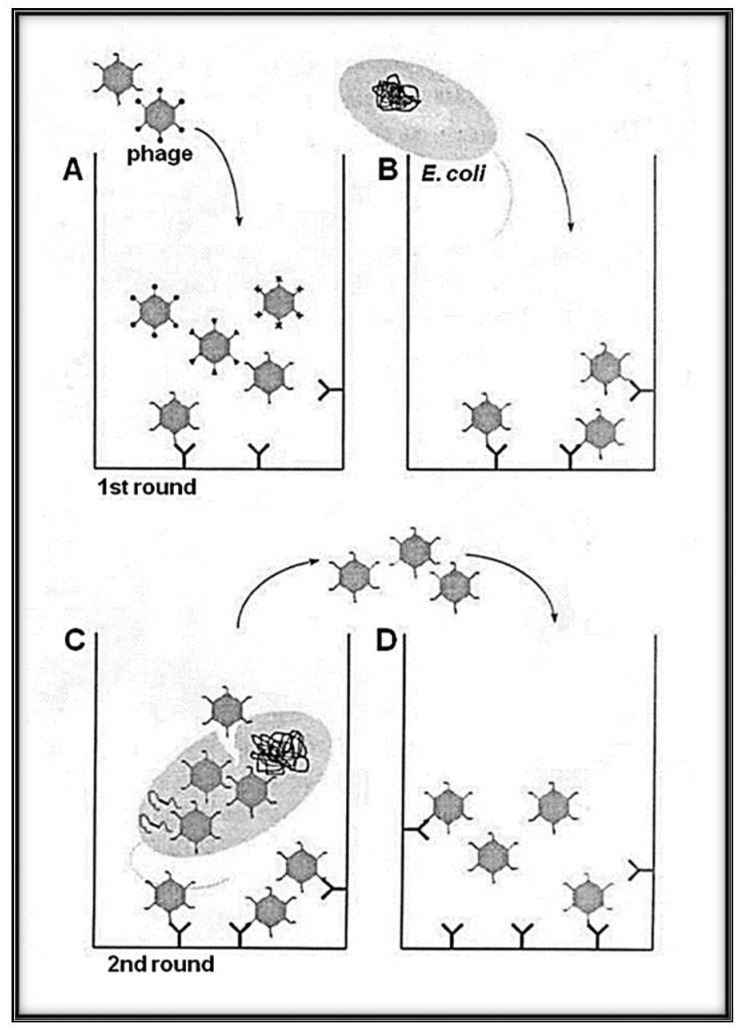
Figure 2.
Schematic representation of the panning methodology with Qβ phage variants (library) with selection and amplification on target avoiding harsh condition (heat and/or acidic elution). (A) A variant of phages solution exposing the library of interest is loaded to the well of a plate pre-coated with the desired target (bio or abio). (B) The indicator bacteria (Escherichia coli Q13) at the log phase growth are added to the well to allow a round of amplification. While the phages, having low affinity to the target, are removed by series of washings, the selected candidates are amplified. (C) High-affinity phages bound to the target can infect E. coli Q13 by adsorbing to the A2 and injecting its RNA via the F+ pilus. (D) Phages newly obtained after indicator E. coli infection were transferred to new wells containing the immobilized target for the next round of biopanning. Several rounds can be used for the enrichment interactions bio-bio or bio-abio.