fig. 5. Efficient Gα subunit folding in a WGE/Ric-8A reconstitution assay is dependent on Ric-8A Ser435 and Thr440 phosphorylation.
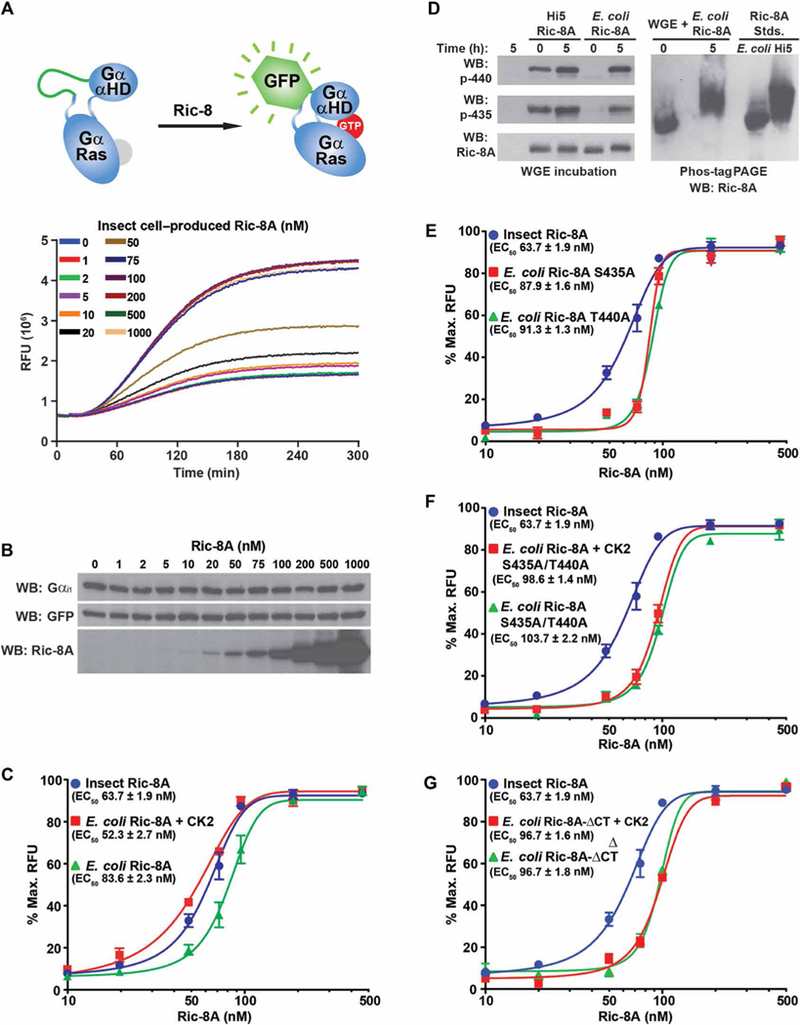
(A) Top: The mRNA encoding a Gαi1 fusion protein with an internal GFP tag was translated in WGE that had been reconstituted with purified rat Ric-8A. Bottom: Folding of the fusion protein was monitored by evolution of GFP fluorescence. (B) Samples for SDS-PAGE were attained at the conclusion of the kinetic Gαi1-GFP translation/folding reactions and analyzed by Western blotting (WB) to detect Gαi1, GFP, and Ric-8A. Data are representative of more than three experiments. (C) Gαi1-GFP mRNA was introduced into WGE translation/ folding reactions reconstituted with the indicated concentrations of insect cell-produced WT Ric-8A or E. coli–produced WT Ric-8A with or without CK2 pretreatment. Maximal Gαi1-GFP relative fluorescence units (RFUs) at 535 nm were plotted versus Ric-8A concentration on semilog plots. The data were fitted to variable Hill slope, four-parameter concentration response functions using the following equation in GraphPad Prism: Y = Ymin + (Ymax – Ymin)/(1 + 10((LogEC50-X)*Hillslope)). EC50 values were estimated from the fitted line functions. Experiments were performed in triplicate, and data are means ± SEM. (D) E. coli–and insect cell–purified Ric-8A proteins were incubated for 5 hours in WGE, resolved by SDS-PAGE and Phos-tag PAGE, and then analyzed by Western blotting. Data are representative of more than three experiments. (E to G) Insect cell–produced WT Ric-8A was used as phosphorylated Ric-8A standard in the WGE/Gαi1-GFP folding assay to compare to the actions of (E) E. coli–produced Ric-8A S435A or T440A, (F) E. coli–produced Ric-8A S435A/T440A treated with or without CK2, or (G) E. coli–produced Ric-8A-ΔCT treated with or without CK2. Data were processed as described in (C). Experiments were performed in triplicate, and data are means ± SEM.
