Fig. 6. Ric-8A-T440 is required for efficient G protein chaperoning activity and signaling in cells.
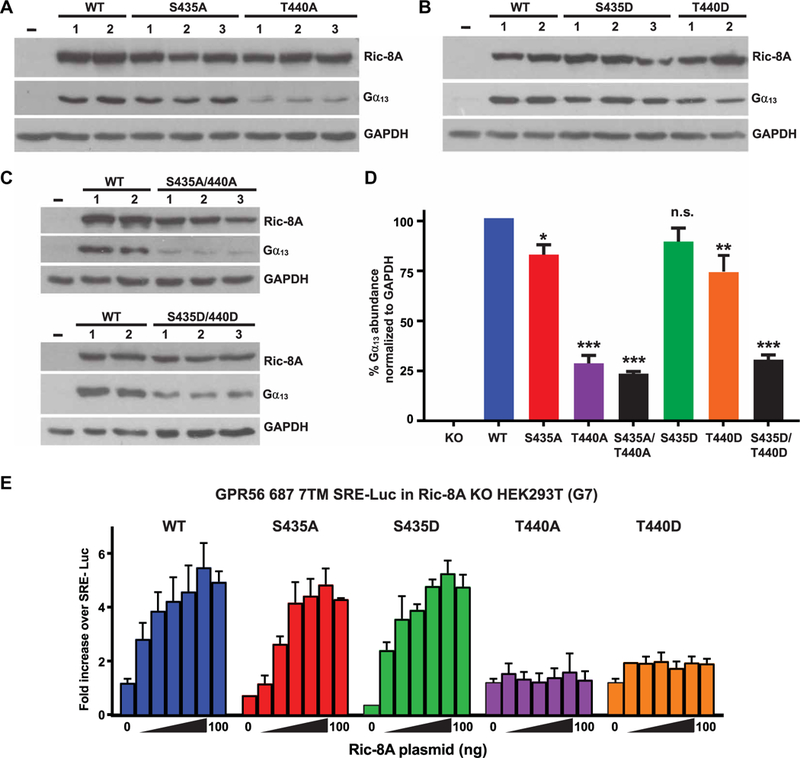
(A to D) A HEK293T cell line (G7) lacking RIC-8A was developed using CRISPR-Cas9 technology. (A to C) Crude membrane preparations from RIC-8A–null cells stably expressing WT or the indicated mutant rat Ric-8A proteins with single alanine or aspartic acid point mutations at the regulatory phosphosites were subjected to quantitative Western blotting for Ric-8A, Gα13, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (D) Relative Gα13 abundances were quantified by pixel densitometry analysis and normalized to the GAPDH signal. Data are means ± SEM of three independent experiments. Statistical significance was determined by one-way analysis of variance (ANOVA), Dunnett’s multiple comparison to WT: *P < 0.05, **P < 0.005, ***P < 0.0001; n.s., not significant. (E) The effects of Ric-8A on GPCR-mediated G13 signaling activity were measured by dual SRE-Luc assay. RIC-8A–null cells were transiently transfected with plasmids expressing constitutively active GPR56, the SRE-Luc reporter, Renilla luciferase, and the indicated amounts of plasmids encoding WT and phosphosite mutant Ric-8A proteins. The accumulated firefly luciferase signal was measured 24 hours after transfection and normalized to the Renilla luciferase signal. Data were normalized to the signal generated from RIC-8A–null cells expressing the luciferase plasmids alone. Data are means ± SEM of three experiments.
