Figure 4: Comparative genome analysis of S. aureus DFU isolates.
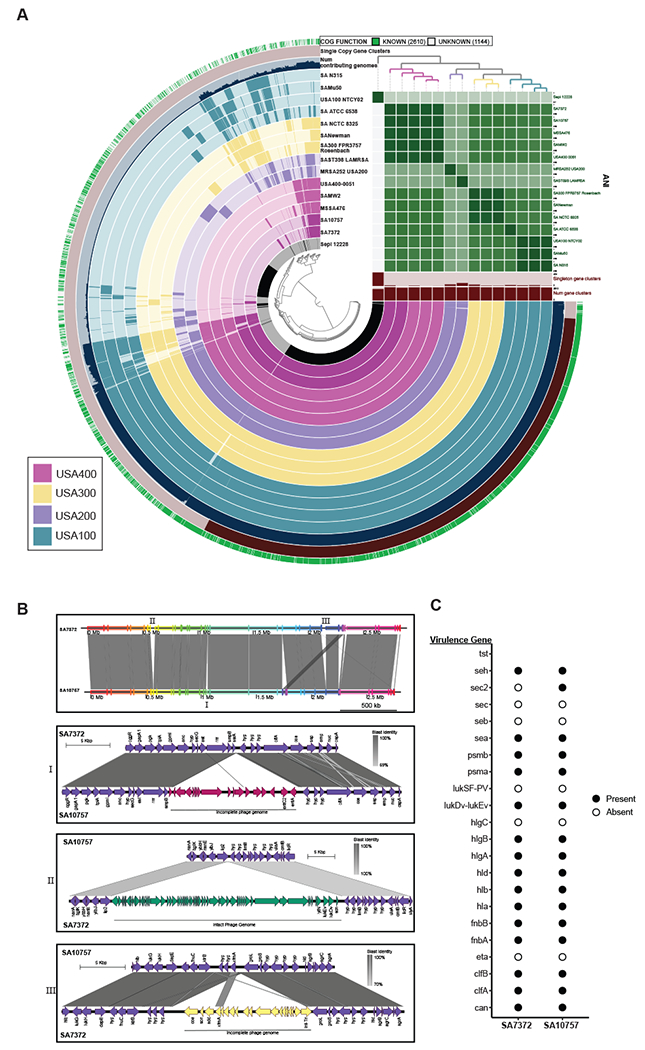
A) Pangenome analysis generated with Anvi’o for 15 S. aureus genomes ordered by gene cluster frequency (opaque=present, transparent=absent). Genomes are colored by monophyletic group. ANI scale 0.95-1 except for S. epidermidis (0.7-1) B) Whole genome and sub-region alignments of SA7372 and SA10757. Homologous blocks are shaded in gray. Phage genomes predicted by PHASTER are denoted with annotation of virulence genes. C) Gene presence (solid) or absence (open) of virulence factors in S. aureus.
