Figure S4.
Phenotypic Abnormality and Individuality of Tumor and Non-tumor Tissue Samples, Related to Figure 4
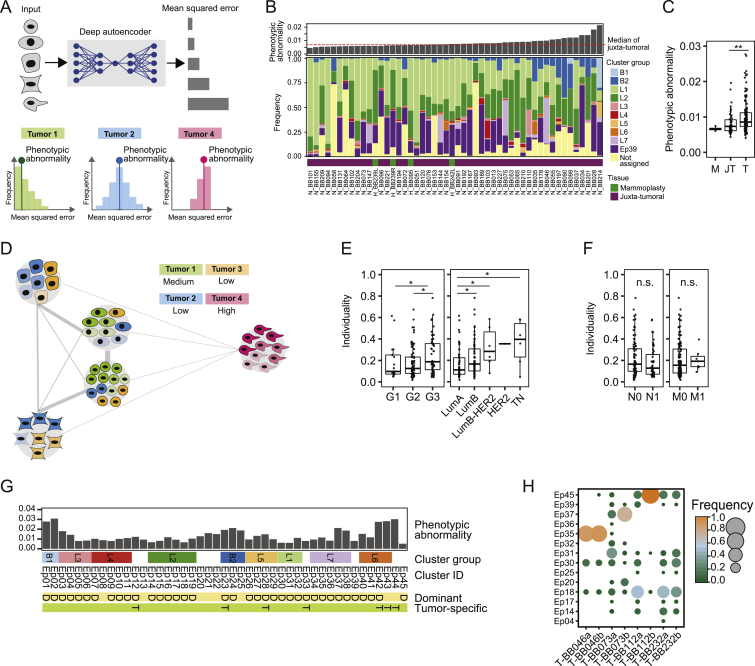
(A) Computation of phenotypic abnormality scores using an autoencoder trained with juxta-tumoral tissue-derived “normal-like” cells. Tumor phenotypic abnormality represents the median Mean Squared Error of all cells of a tumor. (B) Barplot of the phenotypic abnormality scores of mammoplasty and juxta-tumoral tissues and stacked histogram of the frequencies of cells per epithelial cluster group per sample. (C) Phenotypic abnormality scores for mammoplasty (M), juxta-tumoral (JT), and tumor (T) samples. (D) Computation of tumor individuality scores using a k-nearest neighbor graph, where cells of all tumors are grouped based on their phenotype. (E) Tumor individuality scores by grade and subtype. (F) Tumor individuality scores by lymph node status and distant metastasis. (G) Diagram of epithelial clusters that are dominant (D, > 50% of all cells of a sample) or tumor specific (T). (H) Cluster frequency map showing tumors for which two areas of the same tumor were sampled. Wilcoxon rank-sum test was used for statistical analysis. ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001.