Figure 1.
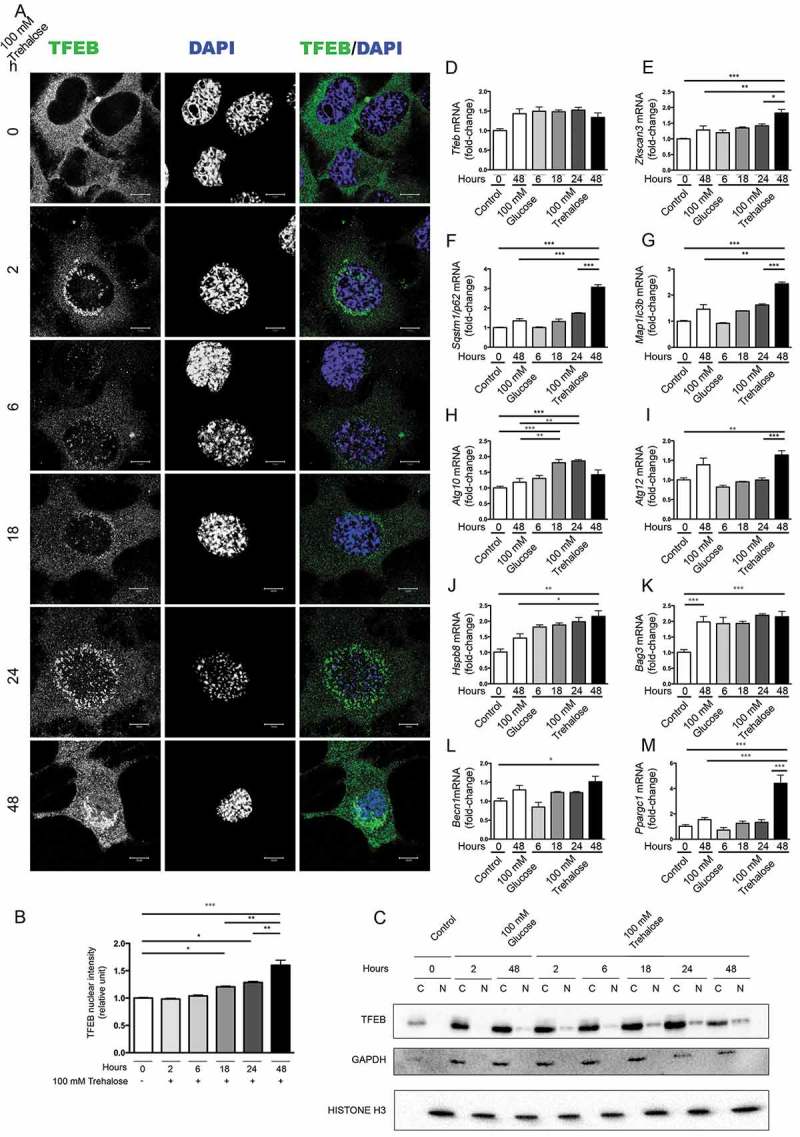
Trehalose activates TFEB nuclear translocation and induces protein quality control genes. (a-m) NSC34 cells were treated with 100 mM trehalose or glucose (as control) for different times. (a) IF analysis performed with anti-TFEB antibody (green), nuclei were stained with DAPI (blue) (63X magnification). Scale bar: 10 μm. (b) The bar graph represents the quantification of TFEB nuclear intensity; the fields were randomly selected and at least 100 cells for each sample were analyzed (n = 3) (*p < 0.05, ** p < 0.005, *** p < 0.001, one-way ANOVA with Tukey’s test). (c) WB analysis of cytoplasmic (C) and nuclear extracts (N). GAPDH and histone H3 were used as loading controls for cytoplasmic and nuclear fractions, respectively. (d-m) RT-qPCR analyses. The relative fold difference in mRNA expression was determined using t = 0 as internal control. Data are means ± SD of 4 independent samples. RT-qPCR on the following mRNA: Tfeb (d); Zkscan3 (e); Sqstm1/p62 (f); Map1lc3b (g); Atg10 (h); Atg12 (i); Hspb8 (j); Bag3 (k); Becn1 (l); Ppargc1A (m). Bar graphs represent the relative fold induction of these genes (*p < 0.05, ** p < 0.005, *** p < 0.001, one-way ANOVA with Tukey’s test).
