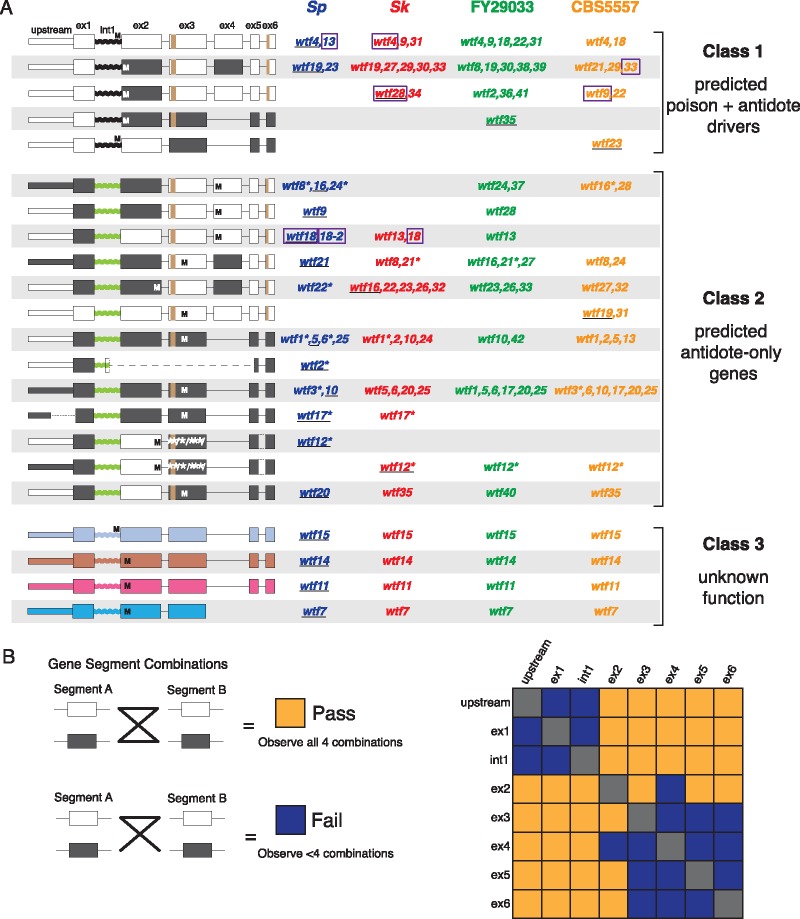
Fig. 2.
Classification of wtf genes based on sequence, and evidence of nonallelic gene conversion. (A) Although they are quite diverged from each other, wtf7, wtf11, wtf14, and wtf15 were placed in a shared class because their sequences are unlike any functionally characterized genes. For the remaining genes, individual gene segments (each exon, intron 1, and the upstream region) from all genes were aligned and classified based on the major clades in maximum likelihood trees (see text for details). Each segment’s clades were color-coded for display purposes (i.e., black/white, black/green coding), and genes were grouped based on gene segment patterns. On the left, we display cartoons of gene structures for each group: boxes indicate exons, “M” indicates in-frame start codons, “/” indicates frameshift mutations, and “*” indicates in-frame stop codons. The repeat regions found in exons 3 and 6 are shown in brown. The names of genes in each class are listed on the right, with the gene illustrated in the cartoon underlined, and pseudogenes denoted with asterisks after gene names. The predicted function of each gene class is shown on the far right. The Class 1 genes are predicted to be meiotic drivers and the Class 2 genes are predicted to be suppressors of drive. Genes with experimentally verified phenotypes have their names outlined with purple boxes. (B) Pairwise four-gamete test for recombination (gene conversion) between all pairs of wtf gene segments for the genes in Classes 1 and 2. Orange boxes indicate that recombination likely occurred because all four segment combinations were observed. Purple boxes indicate that not all segment combinations were observed.