Figure 6.
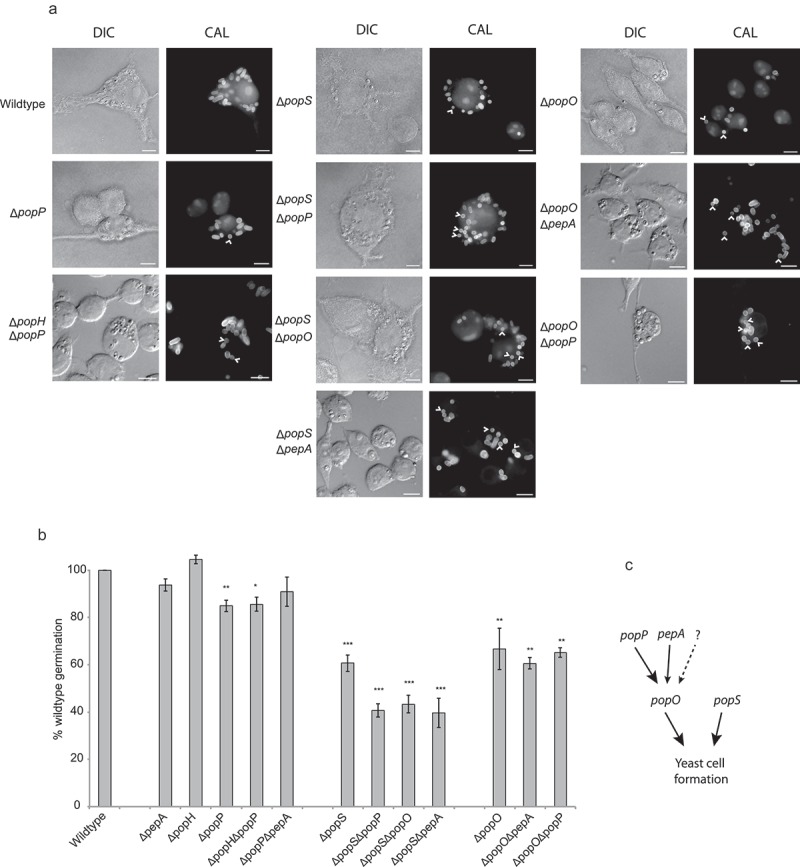
Aspartyl protease-encoding genes are required for appropriate morphogenesis inside host macrophages.
(a) Strains bearing mutations in the pop genes showing yeast cell-specific expression were used to infect murine J774 macrophages and assayed for yeast cell morphogenesis after 24-h post infection. The popP, popO and popS single deletion mutant strains show decreased yeast cell formation in macrophages compared to the wild-type. Double deletion mutant strain combinations for these genes show varying levels of reduction in yeast cell formation with popS combinations showing the greatest change. Representative images using DIC and epifluorescence (CAL, calcofluor staining) are shown. Arrows highlight spores that failed to produce yeast cells. Scale bars = 10 µm. (b) Conversion of dormant conidia into yeast cells (germination) inside J774 macrophages was quantified for each of the mutant strains. Approximately 100 infected J774 macrophage cells infected with each strain in three biological repeat experiments were used to determine germination, and then plotted as a percentage of the wild-type levels. The error bars represent SEM with t-test values falling in the following range * = <0.05, ** = <0.01, *** = <0.001. (c) A model of the postulated genetic interaction pathway based on the severity of their phenotypes and non-additive, epistatic interactions noted for the various combinations of gene deletions and the single mutants. The dotted arrow and question mark denote possible unidentified pathways or proteases of the pop family that may contribute to the formation of yeast cells in T. marneffei during intracellular growth.
