Figure 3.
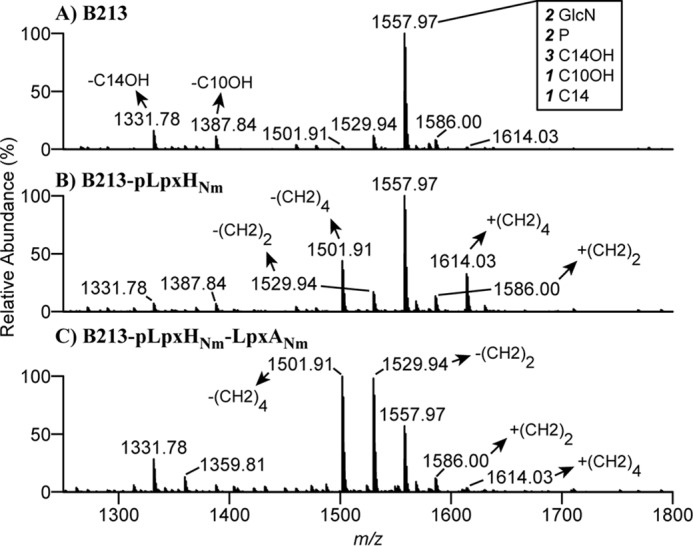
Structural analysis of lipid A. A–C, negative-ion lipid A mass spectra were obtained by in-source CID nano-ESI–FT–MS of intact LPS isolated from cells of B213 (A), B213 expressing lpxHNm (B), and B213 expressing lpxHNm and lpxANm (C). Bacteria were grown for 12 h in Verwey medium in the presence of IPTG. A major singly deprotonated ion at m/z 1557.97 was interpreted as the typical B. pertussis lipid A structure: a diglucosamine (2 GlcN), penta-acylated (three 3OH-C14, one 3OH-C1, and one C14) with two phosphate residues (2 P), as illustrated in Fig. 1B. Additional singly deprotonated lipid A ions were detected in different derivatives, and their interpretations are indicated. Only the m/z range covering lipid A ions is shown.
