Figure 3.
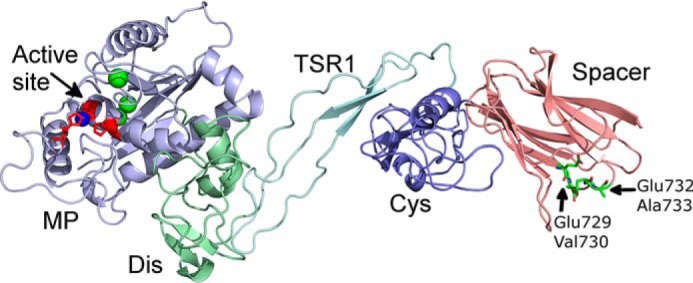
Structural model of ADAMTS7 MP-Spacer domains. The model of the ADAMTS7 Spacer domain structure revealed that the autolytic cleavage sites identified by N-terminal sequencing and TAILS (Glu-729–Val-730 and Glu-732–Ala-733) are present in the surface exposed β3-β4 loop of the Spacer domain. The active-site zinc in the MP domain is shown in blue, structural calcium ions are shown in green. The structure of the N-terminal domains of ADAMTS7 (MP-Spacer) was modeled using the Dis-Spacer structure of ADAMTS13 (3GHM) and ADAMTS1, 4, and 5 MP domain structures (Protein Data bank entries 2RJQ, 2RJP, 4WK7, 2V4B).
