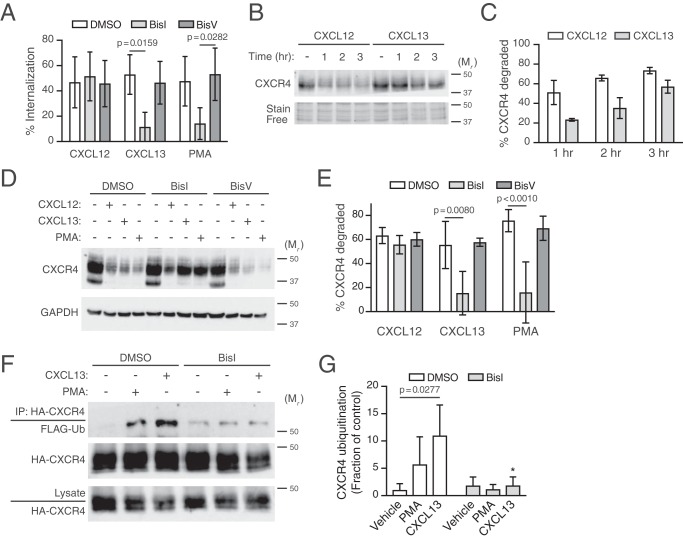
Figure 8.
Role of CXCR5 in cross-regulation of CXCR4. A, CXCR4 internalization was examined in HeLa cells stimulated with 10 nm CXCL12, 100 nm CXCL13, or 10 nm PMA for 5 min. Bars, average receptor internalized from five independent experiments ± S.D. B, CXCR4 degradation was examined in HeLa cells stimulated with 10 nm CXCL12 or 100 nm CXCL13 for the indicated times. Representative CXCR4 immunoblot is shown. Bottom, section of stain-free loading control used for densitometric analysis. The positions of the molecular weight markers are indicated. C, densitometric analysis of CXCR4 levels. Bars, average receptor degraded normalized to a stain-free loading control at each time point relative to vehicle. Error bars, S.D. D, HeLa cells were pretreated with DMSO 10 μm BisI or 10 μm BisV for 30 min, before treatment with 10 nm CXCL12, 100 nm CXCL13, or 10 nm PMA for 3 h. Whole-cell lysates were analyzed by immunoblotting for CXCR4. Bottom, GAPDH immunoblot shown as loading control. Positions of molecular weight markers are indicated. E, CXCR4 degradation was measured by densitometric analysis. Bars, average receptor degraded normalized to GAPDH with each ligand treatment compared with vehicle. Error bars, S.D. The data in A and E were analyzed by two-way ANOVA, followed by Tukey's multiple-comparison test. Adjusted p values are indicated. F, PKC is necessary for CXCL13-promoted ubiquitination of CXCR4. HEK293 cells transfected with FLAG-ubiquitin and HA-CXCR4 were treated with 10 nm PMA or 100 nm CXCL13 for 30 min in the presence or absence of 10 μm BisI. Detection of ubiquitinated CXCR4 was performed as described under “Experimental procedures.” Representative immunoblots are shown. G, CXCR4 ubiquitination was quantified by densitometric analysis. Ubiquitin levels were normalized to CXCR4 levels in immunoprecipitates, and CXCR4 ubiquitination is expressed as a fraction of control or the values from the DMSO and vehicle-treated cells. Bars, average from three independent experiments ± S.D. Data were analyzed by two-way ANOVA, followed by Tukey's post hoc test. The adjusted p value for vehicle:DMSO versus CXCL13:DMSO is indicated. *, adjusted p value 0.0467 for CXCL13:DMSO versus CXCL13:BisI.