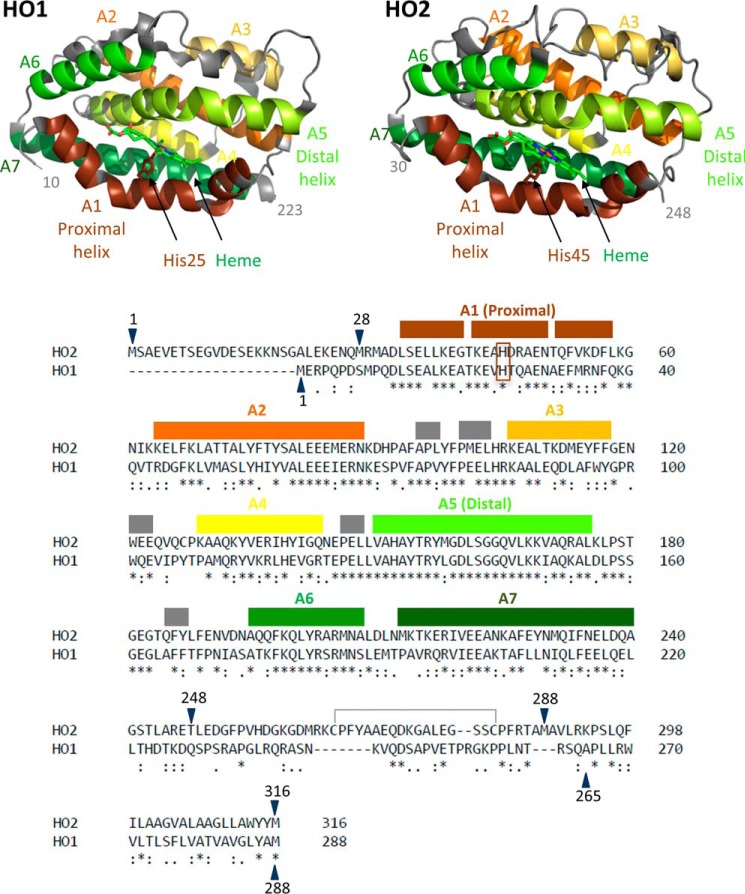
Figure 1.
The structures and sequences of HO1 and HO2. Helices A1–A7 are labeled and indicated by color on the structure of the catalytic core of heme-bound HO1 (Protein Data Bank (PDB) code 1N3U) and the catalytic core of heme-bound HO2 (PDB code 2RGZ). Helices A1–A7, which cover the same indicated regions of both HO2 and HO1, are also indicated using the same color scheme above the linear sequence comparison of HO2 and HO1. An asterisk (*) indicates identical residues, a colon (:) indicates highly similar residues, and a period (.) indicates weakly similar residues. Gray boxes indicate small helical regions that are not numbered. The heme-ligating histidine residues of each protein (His25 in HO1 and His45 in HO2) are in the brown box. The disulfide bond between Cys265 and Cys282 is also indicated. All the constructs studied here that contain the HRMs in the C-terminal tail are oxidized and form a disulfide bond, incapable of binding heme, unless otherwise noted. Constructs of varying amino acid lengths were studied here. Constructs have been truncated on the N-terminal end and/or the C-terminal end as indicated by the blue arrows. A previous alignment differs in the region between HO2 residues 263 and 289 and matches Lys274 (HO2) with Arg254 (HO1) (3).