Figure 11.
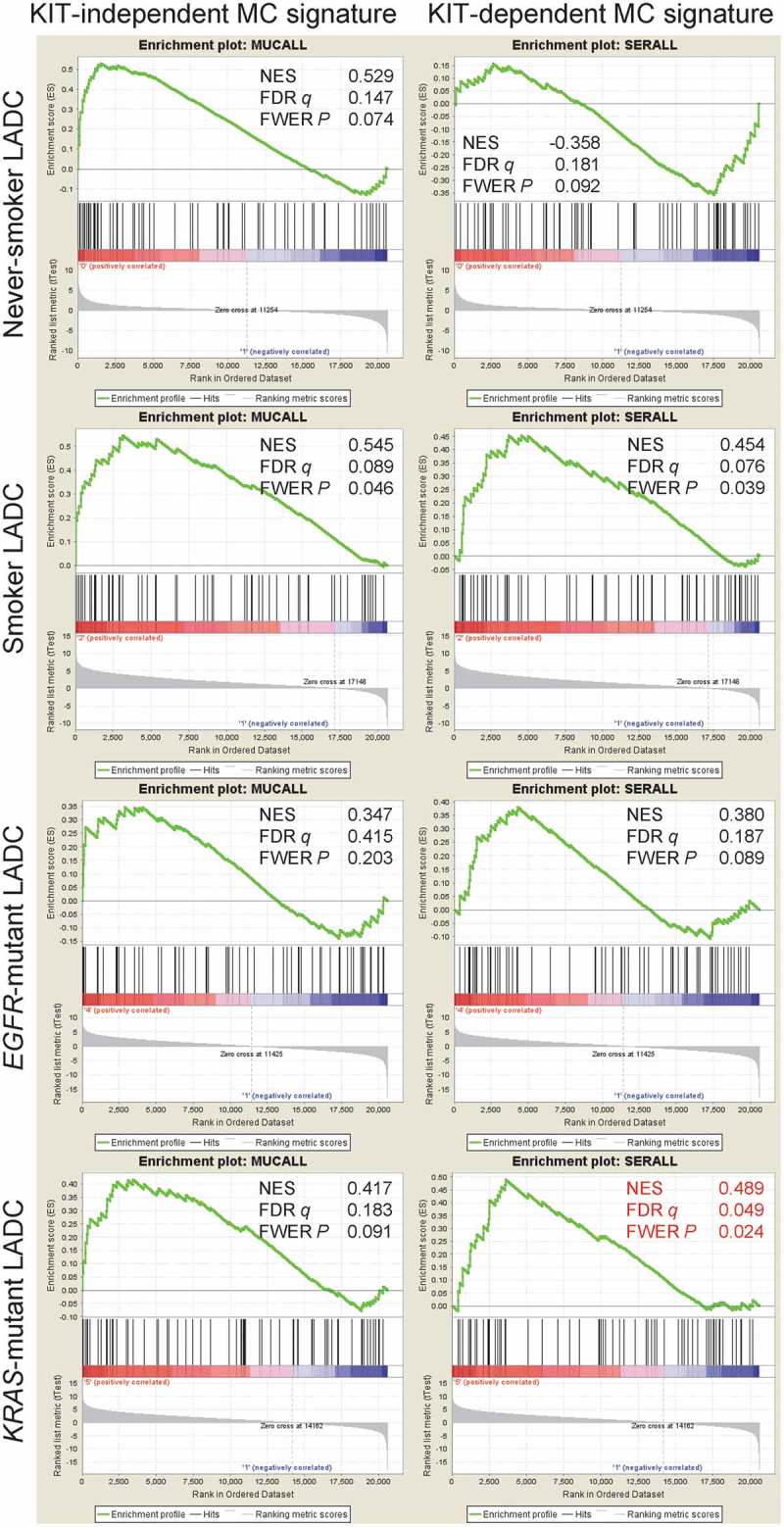
The KIT-dependent mast cell signature is focally enriched in KRAS-mutant lung adenocarcinoma.
Pre-ranked gene set enrichment analysis of the humanized KIT-independent (left) and KIT-dependent (right) mast cell (MC) signatures from Figure 9D against 40 lung adenocarcinoma (LADC) tissues from never-smokers, 40 LADC tissues from smokers, 15 EGFR-mutant LADC, and 22 KRAS-mutant LADC from the Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination (BATTLE) study [Gene Expression Omnibus (GEO) datasets GSE43458 and GSE31852; freely available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43458 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31852].53,55,56 GSEA was performed with the Broad Institute pre-ranked GSEA module software (http://software.broadinstitute.org/gsea/index.jsp).65 Normal lung tissue from 40 never smokers were used as controls (GSE31852). Note that using stringent cut-offs of both false discovery rate (FDR) q values <0.05 and family-wise error rate (FWER) probability (P) values <0.05 the KIT-dependent MC signature is focally enriched in the molecular signature of KRAS-mutant LADC (red fonts). Shown are enrichment plots, normalized enrichment scores (NES), FDR, and FWER values.
