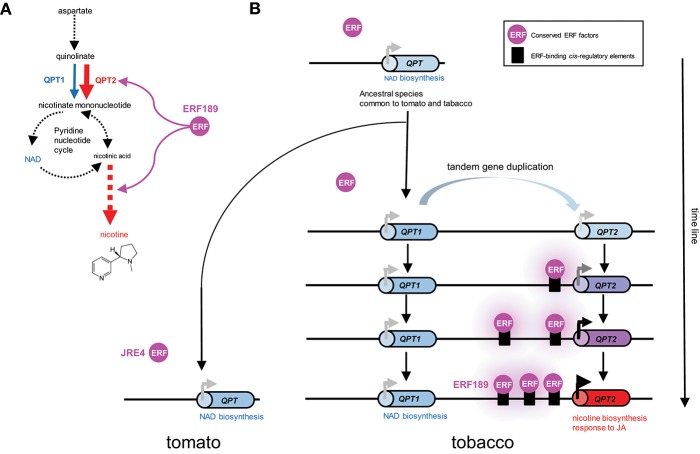
Figure 2.
Recruitment of metabolic genes into regulons under the control of conserved transcription factors through the generation of cognate cis-regulatory elements in their promoter regions; a case study of the evolution of QPT2 in the ERF189-controlled nicotine biosynthesis regulon in the tobacco lineage. (A) Quinolinate phosphoribosyltransferase (QPT) catalyzes a reaction at the entry point in NAD and nicotine biosynthesis. In tobacco, QPT is encoded by distinct QPT1 (blue) and QPT2 (red) genes, which are thought to contribute to NAD and nicotine formation, respectively (Shoji and Hashimoto, 2011a). QPT2 and downstream steps specific to nicotine formation (red arrows) are regulated by ERF189 in tobacco. Steps including multiple enzymes and undefined reactions are represented by broken arrows. (B) Schematic depiction of the evolution of QPT genes in the tomato and tobacco lineages. In the tobacco genome, QPT1 and QPT2, which are thought to have arisen through tandem duplication, are located ~75 kb apart on the chromosome. Tomato contains one QPT gene copy in a genomic region syntenic to the tobacco cluster (Shoji and Hashimoto, 2019). One of the duplicates, QPT2, has become regulated by an evolutionarily conserved ERF transcription factor by gaining ERF-binding cis-regulatory elements in its promoter. Three functional P-box elements bound by ERF189 are present in the proximal promoter region of QPT2 in extant tobacco (Shoji and Hashimoto, 2011a).