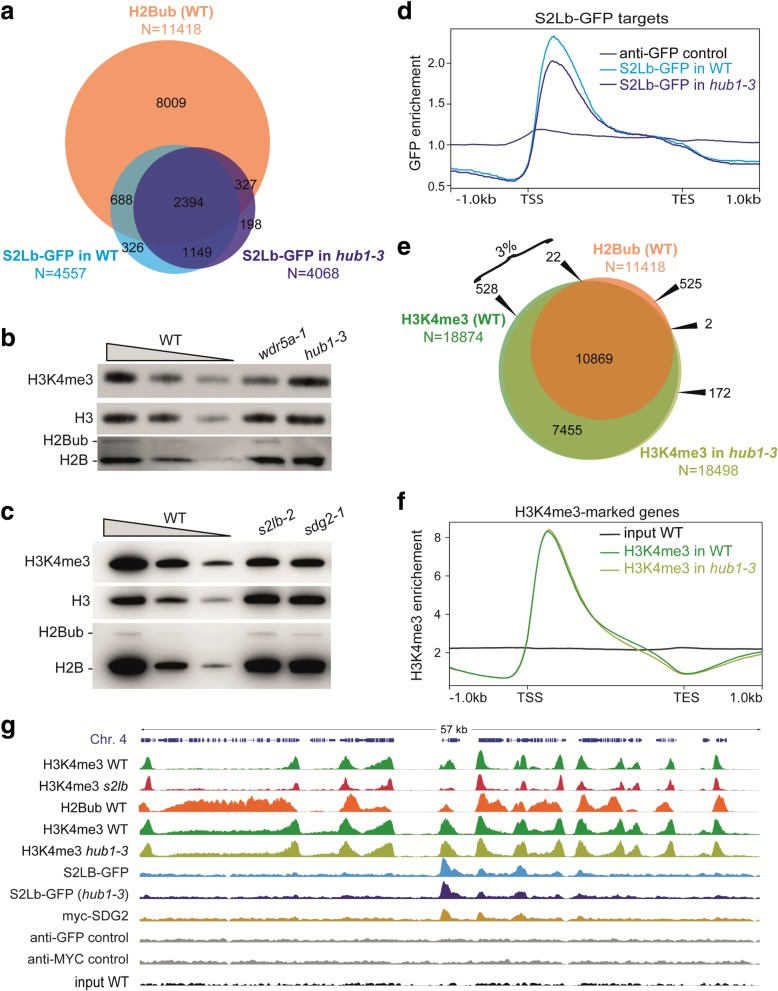
Fig. 6.
S2Lb chromatin association and H3K4me3 deposition are usually independent from histone H2B monoubiquitination. a Overlap between genes marked by H2Bub (from [62]) and by S2LB-GFP in wild-type (WT) or in hub1-3 seedlings. For proper comparison of S2Lb-GFP enrichment, WT/S2Lb::S2Lb-GFP and hub1-3/S2Lb::S2Lb-GFP seedlings were grown and processed for ChIP-seq in parallel. The corresponding gene IDs are listed in Additional file 4: Table S3. b Wild-type H3K4me3 levels in chromatin extracts of HUB loss-of-function plants. Decreasing quantities of chromatin extracts from wild-type plants are shown for comparison, and detection of total histone H3 and H2B forms is used as loading controls. c Wild-type H2Bub levels in S2Lb and SDG2 loss-of-function plants. Immunoblots were performed as in b. d S2Lb-GFP median enrichment in WT/S2Lb::S2Lb-GFP and hub1-3/S2Lb::S2Lb-GFP seedlings on the S2Lb-GFP targeted genes (N = 4557). e Overlap between H2Bub-marked genes (from [62]) and H3K4me3-marked genes in wild-type (WT) or in hub1-3 seedlings (this study). For proper comparison of H3K4me3 marking, wild-type and hub1-3 seedlings were grown and processed for ChIP-seq in parallel, independently from other ChIP-seq in Figs. 3, 4, and 5. The corresponding gene IDs are listed in Additional file 9. f H3K4me3 median enrichment in WT and hub1-3 seedlings on the genes marked by H3K4me3 in WT (N = 18,874). g Genome browser view of H3K4me3, S2Lb-GFP, and MYC-SDG2 over the same region of chromosome 4 shown in Fig. 3. H3K4me3 and GFP or MYC signals in different genetic backgrounds are equally scaled