Figure 3.
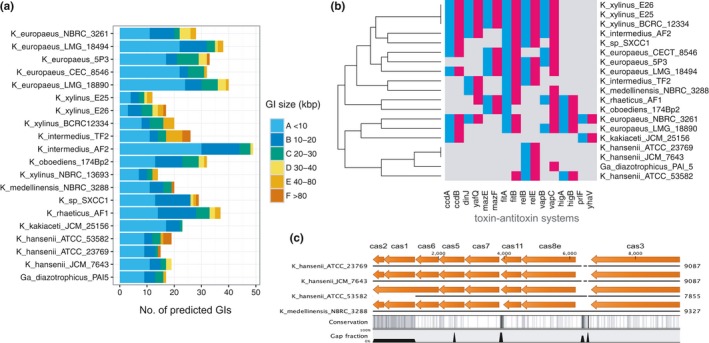
The flexible genome characteristics. (a) Number of predicted GIs of various sizes (kbp) in all analyzed 20 genomes of Komagataeibacter and a Gluconacetobacter diazotrophicus PAl 5 genome. (b) Presence (colored)/absence (gray) map of toxin–antitoxin systems in Komagataeibacter and a Gluconacetobacter strains. Antitoxins are marked in blue, toxins in red and arranged consecutively in pairs. Dendrogram (y‐axis: strains clustering) was generated based on hierarchical clustering analysis. (c) CRISPR‐Cas gene cluster in K. hansenii strains and K. medellinensis NBRC 3288 constituting of eight cas genes. In K. hansenii ATCC 53582, cas1 is incomplete, as it is located at a contig border; cas2 is present on another contig. Figure produced using SnapGene and modified
