Abstract
Objectives
Chemoresistance development represents a major obstacle to the successful treatment of colorectal cancer (CRC). The aim of this study was to elucidate the mechanism by which miR‐506 reverses oxaliplatin chemoresistance in CRC.
Methods
In this study, miR‐506 levels were measured in 74 patients with colon cancer via quantitative real‐time polymerase chain reaction (qRT‐PCR) and in situ hybridization (ISH). We subsequently analysed the relationship between miR‐506 expression and CRC patient survival via the Kaplan‐Meier method. MTT assay demonstrated the fractional survival rates and cell viability of HCT116‐OxR, HCT116‐OxR‐miR‐Ctrl and HCT116‐OxR‐miR‐506 cells treated with oxaliplatin at different concentrations. Cell proliferation and apoptosis were assessed via flow cytometry (FCM) analysis and apoptosis assay. MDR1 mRNA expression and P‐gp protein expression were assessed via qRT‐PCR and Western blotting (WB) respectively. Immunofluorescence (IF) staining demonstrated P‐gp expression in HCT116‐OxR and HCT116‐OxR‐miR‐506 cells. qRT‐PCR and WB were used to detect Wnt/β‐catenin pathway activity after miR‐506 overexpression.
Results
In the present study, in ISH and qRT‐PCR results demonstrated that miR‐506 is weakly expressed in chemoresistant CRC tissues. The low miR‐506 expression group exhibited lower 5‐year OS and lower 5‐year RFS than the high miR‐506 expression group. miR‐506 overexpression inhibited cell growth and increased oxaliplatin‐induced cell apoptosis in HCT116‐OxR cells, as shown via FCM and apoptosis assay. We subsequently noted low MDR1/P‐gp expression in HCT116‐OxR‐miR‐506 cells via qRT‐PCR, WB and IF. Lastly, we demonstrated low MDR1/P‐gp expression in HCT116‐OxR‐miR‐506 cells via inhibition of the Wnt/β‐catenin by WB, MTT and FCM analysis.
Conclusion
Taken together, the findings of our study demonstrate that miR‐506 overexpression in HCT116‐OxR cells enhances oxaliplatin sensitivity by inhibiting MDR1/P‐gp expression via down‐regulation of the Wnt/β‐catenin pathway and thus provide a rationale for the development of miRNA‐based strategies to reverse oxaliplatin resistance in CRC cells.
Keywords: chemoresistance, colorectal cancer, MDR1/P‐gp, miR‐506
1. Introduction
Colorectal cancer (CRC) is the fourth most common cancer and one of the most deadly human malignancies.1 In the last decade, the use of chemotherapeutic agents has significantly increased the survival of patients with advanced CRC. Although vital progress has been made with respect to diagnostic methods, surgical techniques and chemotherapies, resistance to conventional therapies is frequently observed in patients with stage III and IV CRC.
The poor patient prognoses associated with CRC are partially due to resistance to existing therapies; however, our understanding of the mechanisms underlying therapy resistance, as well as the number of available therapeutic options, remains limited. This study found that chemotherapy effectiveness is often limited by abnormal drug resistance‐related protein expression. Common drug resistance proteins include glutathione‐s‐transferase (GST), lung resistance‐related protein (LRP), multi‐drug resistance‐associated protein 1 (MRP1) and permeability‐glycoprotein (P‐gp).2 Overexpression of these proteins results in acquired and/or intrinsic drug resistance in many tumours. However, thus far, the complex mechanisms underlying the effects exerted by these proteins have not been fully elucidated. MicroRNAs (miRNAs) are a class of small, single‐stranded, non‐coding RNA molecules of 19‐24 nucleotides in length3, 4 and have been reported to play an important role in drug resistance in both haematopoietic and solid tumours,5, 6, 7 including CRC.8, 9, 10, 11, 12 Zhou13 reported that miR‐203 induces oxaliplatin resistance in CRC cells by negatively regulating ATM kinase. Wan14 demonstrated that ectopic miR‐320 expression resulted in the inhibition of HCT‐116 cell proliferation and invasion and in hypersensitivity to 5‐Fu and oxaliplatin. miR‐506 is a component of an X chromosome‐linked miRNA cluster that has been reported to function both as an oncogene and as a suppressor of tumour progression.15 In our previous study, we confirmed that the miR‐506‐EZH2 axis modulates CRC invasion and metastasis by inhibiting the Wnt/β‐catenin signalling pathway.16 However, little work has been performed regarding the relationship between miR‐506 and oxaliplatin resistance in chemoresistant CRC cells.
miR‐506 has been identified as a tumour suppressor in CRC; however, the role of miR‐506 in CRC chemoresistance has not been fully studied. In this study, we confirmed that miR‐506 is down‐regulated in chemoresistant CRC cancer tissues, which is associated with a poor prognosis. We also found that miR‐506 inhibited MDR1/P‐gp expression via down‐regulation of the Wnt/β‐catenin pathway to reverse oxaliplatin resistance in HCT116‐OxR cells. These results enhanced our understanding of the molecular mechanisms underlying CRC drug resistance and indicated that miR‐506 may be a therapeutic target in chemoresistant CRC.
2. Materials and methods
2.1. Ethics statement
All patients agreed to participate in the study and provided written informed consent. This study was approved by the ethics board of the Third XiangYa Hospital of Central South University and complied with the Declaration of Helsinki.
2.2. Patient samples
The study enrolled 74 patients with confirmed advanced CRC, including patients diagnosed with stage IV CRC through colonoscopy and magnetic resonance or computed tomography (CT) scan before chemotherapy. Patients ranged from 36‐80 years of age and underwent neoadjuvant chemotherapy (XELOX [capecitabine + oxaliplatin] or mFolFox6 [5‐FU, leucovorin, oxaliplatin]) prior to surgery between 2008 and 2010 at the Department of Gastrointestinal Surgery of the Third Xiangya Hospital of Central South University. Chemotherapy responses were evaluated using the tumour regression grade (TRG) system.17 Patients were divided into two groups based on their response to chemotherapy. The non‐responder (NR) group included TRG1 and TRG2 patients, and the responder (R) group included TRG3 and TRG4 patients. The effects of clinicopathological characteristics, such as age, gender, tumour size, depth of invasion, tumour differentiation, lymph node invasion, TNM stage, metastasis and chemotherapy resistance, on chemotherapy responsiveness were also assessed. Tumours were classified and graded based on the TNM classification advocated by the International Union Against Cancer.
2.3. Cell culture
The human CRC HCT‐116 cells used in this study were purchased from American Type Culture Collection. HCT‐116 cells and HCT116‐OxR cells were cultured in RPMI 1640 medium (Gibco Industries, Inc. Carlsbad, CA, USA), and the medium was supplemented with 10% foetal bovine serum, 100 U/mL penicillin G and 100 μg/mL streptomycin. Oxaliplatin‐resistant HCT‐116 cell (HCT116‐OxR) was established by our laboratory. Briefly, 20 ng/mL of oxaliplatin was used in the beginning to induce drug resistance of HCT‐116 cell line, and thereafter, the concentration of oxaliplatin was increased in gradient. About 7 months later, the cells could stably grow in 20 μg/mL of oxaliplatin, which was named HCT116‐OxR cell line. The HCT116‐OxR cells were seeded in the medium additionally contained 5 μg/mL oxaliplatin, so as to maintain the drug‐resistant phenotype. Both cell lines were incubated in 5% CO2 at 37°C in 100% humidity.
2.4. In situ hybridization analysis
In situ hybridization (ISH) analysis was performed according to a previously described method.18 Antisense oligonucleotide probes for miR‐506 (Exiqon Inc., Woburn, MA, USA) were used for ISH.
2.5. Immunofluorescence staining
For immunofluorescence (IF), cells were seeded on cover slips in 24‐well plates overnight and then fixed in 4% paraformaldehyde in phosphate‐buffered saline (PBS) for 10 minutes, washed twice with PBS, and then permeabilized with 0.1% Triton X‐100 in PBS for 10 minutes. Fixed cells were pre‐incubated in PBS containing 5% BSA for 30 minutes at room temperature. The cells were stained with primary antibody (anti‐P‐gp monoclonal antibody, 1:200 dilutions) for 1 hour at room temperature, followed by incubation with secondary antibody conjugated with FITC. DAPI (0.1 μg/mL) was added to the secondary antibody mixture to visualize nuclei. Fluorescence images were collected and analysed using an inverted fluorescence microscope.
2.6. Cell cycle analysis
At 48 hours after transfection, the cells were harvested, washed with PBS solution and fixed in 70% ethanol overnight at 4°C. After fixation, the cells were washed twice with PBS before incubation in propidium iodide (PI)/RNase A solution (5 μg/mL PI and 100 mg/mL RNase A) at room temperature in the dark for 30 minutes. Stained cells were analysed using a FACSCalibur flow cytometer (Becton‐Dickinson, San Jose, CA, USA), and the analysis was completed within 30 minutes.
2.7. Apoptosis analysis
Cell apoptosis analysis was performed using a Phycoerythrin (PE)‐AnnexinV Apoptosis Detection Kit (BD PharMingen, San Jose, CA, USA). For cell apoptosis analysis, cells were seeded in six‐well plates at a density of 8 × 105 per well. Twenty‐four hours after transfection, oxaliplatin was added at a final concentration of 5 μg/mL. Forty‐eight hours after oxaliplatin treatment, adherent HCT116‐OxR cells in the suspension were harvested and labelled with AnnexinV for 15 minutes in the dark. Then, 50 μg/mL PI was added to each sample before the apoptotic cell distribution was analysed via flow cytometry (FCM) (BD LSRII, San Jose, CA, USA). The experiments were repeated three times.
2.8. MTT assay and drug sensitivity assay
HCT116‐OxR, HCT116‐OxR‐miR‐ctrl and HCT116‐OxR‐miR‐506 CRC cells were seeded in 96‐well plates at a density of 3000/well in 100 μL of medium and incubated overnight to allow attachment. The next day, 100 μL of a stock solution of oxaliplatin was added to the cell suspension at a 2X final concentration. After incubation for 72 hours at 37°C, 40 μL of MTT (3‐(4, 5‐dimethylthiazol‐2‐yl)‐2, 5‐diphenyltetrazolium bromide; 3 mg/mL) was added to each well, and the cells were incubated for an additional 24 hours. After the supernatant was removed, the formazan precipitates in the cells were dissolved in 150 μL of dimethyl sulfoxide. Absorbance was measured with a microplate reader at a wavelength of 570 nm. The chemosensitivity of oxaliplatin was expressed as IC50 (concentration inducing 50% cytotoxicity). After transfection, the HCT116‐OxR, HCT116‐OxR‐miR‐506 and HCT116‐OxR‐miR‐506‐SB‐216763 cells were seeded into 96‐well plates, and oxaliplatin was added at the following concentrations: 6.25, 12.5, 25, 50 and 100 μg/mL. The concentration of SB‐216763 is 5 μmol/L.19 Cell viability was measured via MTT assay.
2.9. Western blot
Total proteins were harvested from cultured cells or fresh frozen tissues and lysed by ice‐cold lysis buffer. Proteins were separated via 6% SDS‐polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% non‐fat milk in PBS for 1 hour at room temperature and then immunoblotted with the following primary antibodies: rabbit‐anti‐human P‐gp, MRP1, LRP, GST (1:1000; Affinity biosciences Inc, OH, USA) and β‐actin (1:800; Affinity biosciences Inc, OH, USA), followed by incubation with horseradish peroxidase‐conjugated secondary antibodies (1:5000; Bioworld Technology, Inc, NanJing, China) for 1.5 hours at room temperature. Reactive bands were visualized using an enhanced chemiluminescence system. Band intensity was quantified using an image analysis system (Quantity One v4.62, Bio‐Rad Laboratories Co., Ltd. Hercules, CA, United States). The experiment was repeated three times.
2.10. Quantitative RT‐PCR
Total RNA was extracted from cells or tissues using TRIzol (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. For mature miR‐506 detection, total RNA was polyadenylated using poly(A) polymerase (Ambion, Austin, TX, USA), as described previously. Reverse transcription was performed using poly(A)‐tailed total RNA, a reverse transcription primer and ImPro‐II Reverse Transcriptase (Promega, Madison, WI, USA), according to the manufacturer's instructions. Quantitative real‐time PCR (qRT‐PCR) was performed as described in the instructions provided with the Fast Start Universal SYBR Green Master Mix (Rox) (Roche Diagnostics GmbH, Mannheim, Germany). U6 was used as an internal control; other specific primers were purchased from Invitrogen.
2.11. miRNA transfection
The miRNA‐506 mimic and negative control were obtained from GenePharma (Shanghai, China). The sequence of the miR‐506 mimic was 5′‐UAAGGCACCCUUCUGAGUAGA‐3′. Cells (5×105 cells/2 mL/well) were seeded in six‐well plates at 60% confluence. After 48 hours, the miRNA‐506 mimic or the negative control was transfected into the cells using Lipofectamine 2000 (Invitrogen) at a final concentration of 50 nmol/L, according to the manufacturer's instructions.
2.12. Statistical analysis
All values are expressed as the mean ± standard deviation (SD). The significance of the differences was determined via one‐way ANOVA or Student's t‐test. An χ2 test was used to evaluate the relationship between expression and patient clinicopathological characteristics. The Kaplan‐Meier method was employed for survival analysis, and the differences in survival probability were estimated using the log‐rank test by Prism software (GraphPad Software 6). P<.05 was considered statistically significant. Statistical analysis was performed using spss version 20.0 (SPSS, Inc., Chicago, IL, USA).
3. Results
3.1. miR‐506 expression was down‐regulated in chemoresistant CRC cancer tissues and was correlated with survival
To study the expression levels of miR‐506 in chemotherapy‐treated CRC tissues, mature miR‐506 was analysed in 31 NR CRC tissue samples and 43 R CRC tissue samples via qRT‐PCR. The expression of miR‐506 is no difference between the pre‐chemotherapy and after chemotherapy in the NR CRC tissue samples (Figure 1A). In the R CRC tissue samples, the expression of miR‐506 is higher in the pre‐chemotherapy than after chemotherapy (Figure 1B). ISH analysis showed that miR‐506 expression was significantly down‐regulated in NR CRC tissue samples compared to R CRC tissue samples (P<.05, Figure 1D), and the qRT‐PCR results indicated that miR‐506 is down‐regulated in chemoresistant CRC (P<.05, Figure 1C). We further analysed the relationship between the miR‐506 expression levels and the clinicopathological characteristics of CRC patients (Table 1). The patients were stratified into two groups based on the median miR‐506 expression levels; the miR‐506 levels were negatively associated with tumour size (P=.00015), lymph node invasion (P=.004), TNM stage (P=.009), CEA (P=.010), CA199 (P=.018) and chemotherapy resistant (P=.00041). We conducted 5‐year follow‐ups of these patients. The 5‐year overall survival (OS) rates and 5‐year relapse‐free survival (RFS) rates was 30.80% and 13.51% in the low miR‐506 expression group and 67.04% and 59.33% in the high miR‐506 expression group. Kaplan‐Meier analysis revealed that patients with low miR‐506 expression were found to have significantly worse 5‐year OS and 5‐year RFS rates (P<.05).
Figure 1.
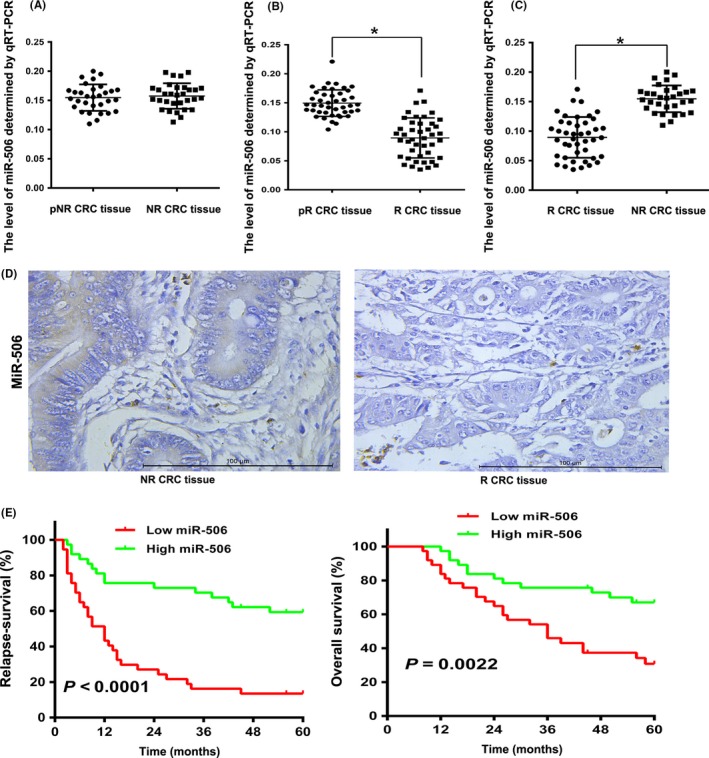
miR‐506 expression was down‐regulated in chemoresistant colorectal cancer (CRC) tissues and correlated with survival. A, The relative expression levels of miR‐506 were assessed via qRT‐PCR between pre‐chemotherapy (pNR CRC) and after chemotherapy in 31 non‐responder colorectal cancer (NR CRC) tissues. B, The relative expression levels of miR‐506 were assessed via qRT‐PCR between pre‐chemotherapy (pR CRC) and after chemotherapy in 43 responder colorectal cancer (R CRC) tissues. C, The relative expression levels of miR‐506 were assessed via qRT‐PCR in 43 R CRC tissues and 31 NR CRC tissues. D, Analysis of miR‐506 expression in chemoresistant CRC cancer tissues and non‐chemoresistant CRC cancer tissues by in situ hybridization (ISH) (×400). E, Kaplan‐Meier analysis revealed that the OS and RFS rates of the high miR‐506 expression group was higher than those of the low miR‐506 expression group (*P<.05)
Table 1.
Relationship between miRNA‐506 and clinicopathological parameters in 74 colorectal cancer patients
| Variable | All cases | miR‐506 expression | χ2 | P a | |
|---|---|---|---|---|---|
| High | Low | ||||
| Ages (years) | |||||
| <60 | 25 | 10 | 15 | 1.510 | 0.219 |
| ≥60 | 49 | 27 | 22 | ||
| Gender | |||||
| Male | 57 | 25 | 32 | 3.742 | 0.053 |
| Female | 17 | 12 | 5 | ||
| Lymph node invasion | |||||
| N1 | 30 | 21 | 9 | 8.073 | 0.004 |
| N2 | 44 | 16 | 28 | ||
| TMN stage | |||||
| III | 45 | 28 | 17 | 6.861 | 0.009 |
| IV | 29 | 9 | 20 | ||
| CA‐199 | |||||
| <37 | 30 | 20 | 10 | 5.606 | 0.018 |
| ≥37 | 44 | 17 | 27 | ||
| CEA | |||||
| <5 | 39 | 25 | 14 | 6.560 | 0.010 |
| ≥5 | 35 | 12 | 23 | ||
| Tumour size | |||||
| <5 | 30 | 23 | 7 | 14.351 | <0.001 |
| ≥5 | 44 | 14 | 30 | ||
| Chemotherapy resistant | |||||
| NR | 31 | 23 | 8 | 12.491 | <0.001 |
| R | 43 | 14 | 29 | ||
Probability, P, from χ2 test.
3.2. Ectopic miR‐506 expression enhanced oxaliplatin sensitivity in chemoresistant CRC cells
HCT‐116 and HCT116‐OxR cells were treated with different concentrations of oxaliplatin (0‐100 μg/mL) for 48 hours. The IC50 value of oxaliplatin was 9.18 and 70.01 μg/mL in the parental cell line HCT‐116 and in the HCT116‐OxR cell line. The resistance index (RI), the ratio of the IC50 of oxaliplatin in HCT116‐OxR cells to that of the parental HCT‐116 cells, was 7.63 (Figure 2A). As the RI value of the cells was >3, the HCT116‐OxR was considered to display chemoresistant characteristics. First, we compared miR‐506 expression levels in parental CRC HCT‐116 cells and oxaliplatin‐resistant CRC HCT116‐OxR cells. We confirmed the low expression levels of miR‐506 via qRT‐PCR in the chemoresistant cell line compared with the parental cell line (Figure 2B). To determine the potential role of miR‐506 in CRC chemoresistance, HCT116‐OxR cells were infected with miR‐506 mimic to overexpress miR‐506. Transfection efficiency was confirmed via qRT‐PCR (Figure 2C). Then, we examined the fractional survival rates of HCT116‐OxR (Blank), HCT116‐OxR‐miR‐Ctrl (miR‐Ctrl) and HCT116‐OxR‐miR‐506 (miR‐506) cells, which were treated with oxaliplatin at different concentrations, via MTT assay. The IC50 of oxaliplatin in HCT116‐OxR‐miR‐Ctrl and HCT116‐OxR‐miR‐506 cells was 41.54 and 14.75 μg/mL. The RI of oxaliplatin in HCT116‐OxR‐miR‐Ctrl cells and HCT116‐OxR‐miR‐506 to that of the parental HCT116 cells was 4.52 and 1.61, the HCT116‐OxR‐miR‐ctrl was considered to display chemoresistant characteristics and HCT116‐OxR‐miR‐506 was considered as non‐chemoresistant characteristics. The results suggested that miR‐506 overexpression resensitized HCT116‐OxR cells to oxaliplatin treatment, as evidenced by the leftward shift of the growth inhibition curve (Figure 2D). The fractional survival rates of the three groups were most significant at 5 μg/mL oxaliplatin. Decreased cancer cell proliferation is closely related to cell cycle arrest. Thus, we performed FCM analyses to evaluate the effects of miR‐506 overexpression on the cell cycle under 5 μg/mL oxaliplatin treatment. miR‐506 significantly increased the number of cells in G1 phase and elicited corresponding reductions in the percentages of HCT116‐OxR cells in S and G2 phase (Figure 2E), indicating that miR‐506 suppresses HCT116‐OxR cell growth. Lastly, the ability of miR‐506 to induce apoptosis in HCT116‐OxR cells was evaluated via co‐staining with AnnexinV and PI. We found that miR‐506 overexpression significantly enhanced 5 μg/mL oxaliplatin‐induced apoptosis in HCT116‐OxR cells compared with other two groups, resulting in a decrease in the viable cell population (Figure 2F). Briefly, miR‐506 up‐regulation resensitized HCT116‐OxR cells to oxaliplatin treatment.
Figure 2.
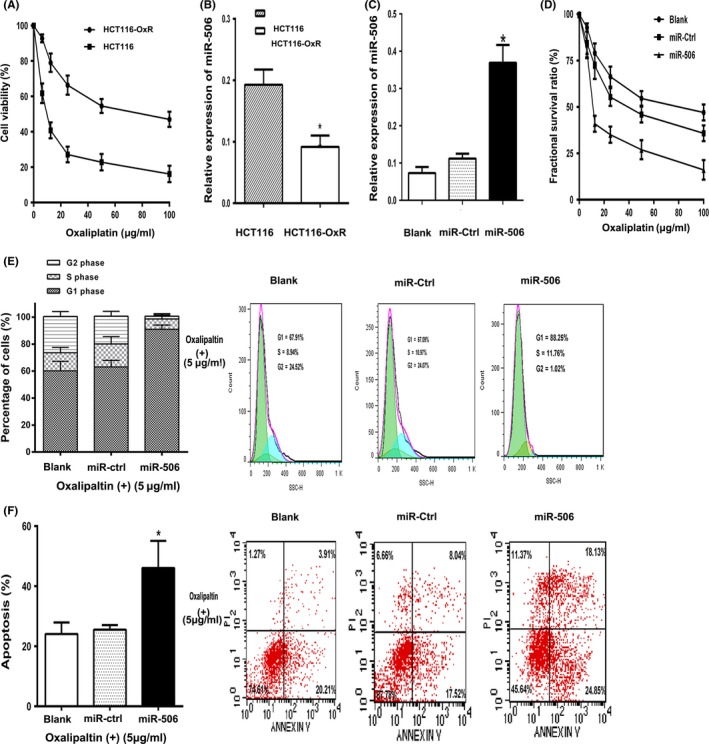
The ectopic expression of miR‐506 inhibited the proliferation of oxaliplatin‐resistant colorectal cancer cells. A, The cell viability of HCT116 and HCT116‐OxR were treated with different concentrations of oxaliplatin for 48 h. B, miR‐506 was down‐regulated in HCT116‐OxR cells. C, miR‐506 was up‐regulated in HCT116‐OxR cells via the transfection of a miR‐506 mimic. After 48 h, the level of miR‐506 was detected via qRT‐PCR. D, Overexpression of miR‐506 significantly decreased the growth‐inhibitory effect of oxaliplatin in HCT116‐OxR cells, as measured by MTT assay. E, The effect of miR‐506 on the cell cycle distribution of HCT116‐OxR was monitored via flow cytometry at 5 μg/mL oxaliplatin concentration. The miR‐506‐overexpression HCT116‐OxR cells were arrested at G1 phase of the cell cycle, resulting in a corresponding reducing in the percentage of cells in S and G2/M phases. F, The proportion of annexin V‐positive apoptotic cells was evaluated by flow cytometry using annexin V allophycocyanin and propidium iodide staining of HCT116‐OxR after transfection of a miR‐506 mimic. HCT116‐OxR cells were used as blank group and HCT116‐OxR‐miR‐Ctrl cells were used as negative control. The data are shown as the means ± SD of three replicates (*P<.05)
3.3. Correlation between resistance‐related gene expression and protein and miR‐506 levels in HCT116‐OxR cells
To elucidate the mechanism underlying the miR‐506‐mediated reversal of oxaliplatin resistance in CRC, the gene and relative protein expression levels of MDR1, LRP, GST and MRP1 in miR‐506‐transfected HCT116‐OxR cells were evaluated via qRT‐PCR and Western blotting (WB) (Figure 3A,B). MDR1/P‐gp expression was lower in HCT116‐OxR‐miR‐506 cells than in HCT116‐OxR and HCT116‐OxR‐Ctrl cells (P<.05). A similar result was observed via IF staining (Figure 3C). These findings indicate that miR‐506 strengthens the sensitivity of HCT116‐OxR cells to oxaliplatin by inhibiting P‐gp expression.
Figure 3.
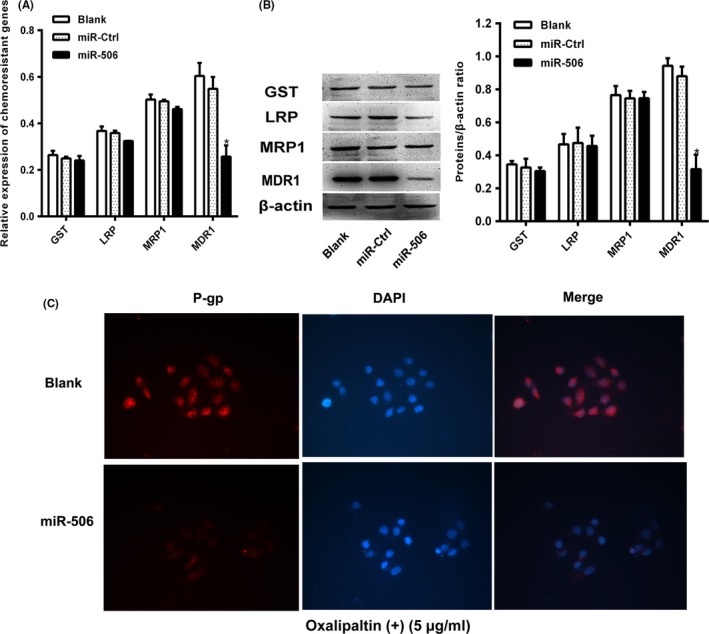
miR‐506 down‐regulated MDR1/P‐gp expression in HCT116‐OxR. A, The mRNA level of MDR1 was decreased after transfection with the miR‐506 mimic of the relative chemoresistant genes as demonstrated by qRT‐PCR. B, The protein level of MDR1 was decreased after transfection with the miR‐506 mimic of the relative chemoresistant proteins as demonstrated by Western blot. C, Expression of P‐gp detected by immunofluorescence staining.HCT116‐OxR‐miR‐506 cells showed low levels of fluorescent staining of P‐gp, whereas maximal staining of P‐gp was observed in HCT116‐OxR cells, readily distinguished from background. Zoom: 200×. *P<.05)
3.4. miR‐506 up‐regulation in HCT116‐OxR cells inhibited MDR1/P‐gp expression via the Wnt/β‐catenin pathway
Studies have revealed that the Wnt/β‐catenin signalling pathway plays an important role in cell proliferation, differentiation, invasion, migration and chemoresistance in many types of cancers. Our previous study showed that miR‐506 acts as a negative regulator of β‐catenin to inhibit the Wnt/β‐catenin pathway. Shen20 and Zhang21 showed that inhibition of Wnt/β‐catenin signalling down‐regulates P‐gp and reverses multi‐drug resistance in cholangiocarcinoma and ovarian cancer. Therefore, we speculated that miR‐506 may play an important role in the inhibition of MDR1/P‐gp expression by down‐regulating Wnt/β‐catenin signalling pathway activity. To validate this inference, we used qRT‐PCR and WB to detect Wnt/β‐catenin pathway activity after miR‐506 overexpression in HCT‐116 and HCT116‐OxR cell. The expression level of MDR1, cyclinD1 and β‐catenin were decreased in HCT‐116 compared with HCT116‐OxR, as demonstrated by qRT‐PCR and WB [S1]. And we detected low β‐catenin expression and low cyclinD1 expression in HCT116‐OxR‐miR‐506 cells, indicating that the Wnt/β‐catenin pathway was inhibited. Then, we added the Wnt/β‐catenin activator SB‐216763 (5 μmol/L) and noted parallel increases in β‐catenin, cyclinD1 and P‐gp expression in HCT116‐OxR‐miR‐506‐SB‐216763 cells (Figure 4A). Next, we noted that fractional survival ratio was apparently lower in the HCT116‐OxR‐miR‐506 group than in the other two groups under different oxaliplatin concentrations (Figure 4B). The IC50 of oxaliplatin in HCT116‐OxR‐miR‐506‐SB216763 cells was 45.34 μg/mL. The RI of oxaliplatin in HCT116‐OxR‐miR‐506‐SB216763 cells to that of the parental HCT‐116 cells was 4.94. The HCT116‐OxR‐miR‐506‐SB216763 was considered to display chemoresistant characteristics. In addition, we detected changes in cell proliferation and apoptosis after activation of the Wnt/β‐catenin pathway. Cell proliferation and apoptosis were significantly restored after activation of the Wnt/β‐catenin pathway (Figure 4C). Taken together, these data indicate that miR‐506 up‐regulation decreases MDR1/P‐gp expression in HCT116‐OxR cells by inhibiting the Wnt/β‐catenin pathway to enhance the sensitivity of human CRC cells to oxaliplatin.
Figure 4.
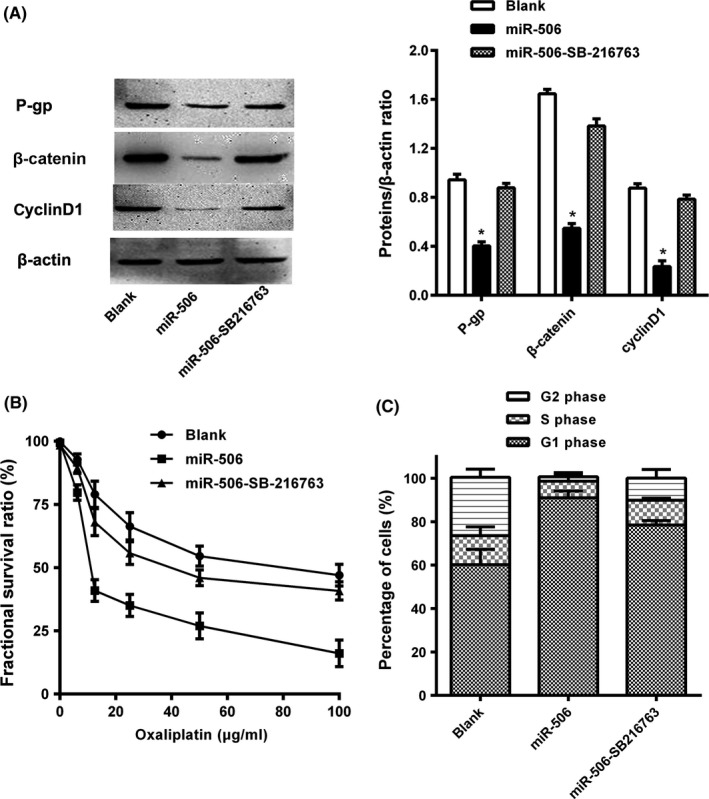
miR‐506 regulated the P‐gp expression via modulated the Wnt/β‐catenin pathway. A, The protein levels of P‐gp, cyclinD1 and β‐catenin were decreased after transfection with the miR‐506 mimic and then the P‐gp, cyclinD1 and β‐catenin in HCT116‐OxR‐miR‐506 cells were increased after SB‐216763 treatment for 6 h, as demonstrated by Western blot. B, Overexpression of miR‐506 significantly decreased the growth‐inhibitory effect of oxaliplatin in HCT116‐OxR cells, while GSK‐3β inhibitor (SB‐216763) increased the growth‐inhibitory effect of oxaliplatin in HCT116‐OxR‐miR‐506 cells as measured by MTT assay. C, The effect of miR‐506 and SB‐216763 on the cell cycle distribution of HCT116‐OxR was monitored via flow cytometry. The HCT116‐OxR‐miR‐506‐SB‐216763 cells which arrested at G1 phase of the cell cycle were less than the HCT116‐OxR‐miR‐506 cells. The data are shown as the means ± SD of three replicates (*P<.05)
4. Discussion
Therapeutic resistance is a major challenge in effective cancer treatment. Oxaliplatin resistance is currently one of the major obstacles in CRC chemotherapy. Although great progress has been made in cancer therapy, most clinically effective chemotherapy inducers are ineffective due to the complexity of the mechanisms underlying chemotherapy resistance. CRC cells develop resistance to oxaliplatin through reduced cellular uptake, impaired DNA adduct formation, DNA repair gene alternations and transporter modifications. MDR1 overexpression has rendered many currently available chemotherapeutic agents ineffective. In clinical situations, MDR1 overexpression often results in P‐gp overexpression in tumour cell membranes. This protein, encoded by MDR1, is the most important membrane transporter for preventing intracellular accumulation of anti‐cancer drugs. Many agents that modulate the function of P‐gp have been identified, including calcium channel blockers, calmodulin antagonists, steroidal agents, protein kinase C inhibitors, immunosuppressive drugs, antibiotics and surfactants.22 Improving chemosensitivity by targeting P‐gp has been extensively used as a strategy for cancer therapy. A few studies have used oxaliplatin‐resistant CRC cells as a model. Montazami23 used SW480‐OxR colon cancer cells as a model to demonstrate that siRNA‐mediated silencing of MDR1 can reverse oxaliplatin resistance. In this study, we used HCT116‐OxR CRC cells treated with oxaliplatin. Our results showed that reducing P‐gp expression in HCT116‐OxR CRC cells resulted in loss of the chemoresistant phenotype in HCT116‐OxR CRC cells, which we confirmed at both the RNA and protein levels using RT‐PCR and WB. Aberrant miRNA expression has recently been implicated in the development of chemoresistance in CRC; however, the molecular mechanisms underlying the regulation of MDR1/P‐gp by miRNA have not been elucidated.
Several reports have partially described the mechanisms by which miR‐506 enhances or reduces drug resistance. Liu24, 25 found that miR‐506 augments chemotherapy responses through RAD51 regulation in serous ovarian cancers. Tong12 reported that miR‐506 overexpression in HCPT‐resistant colon cancer SW1116/HCPT cells confers resistance to HCPT by inhibiting PPARα expression. However, the mechanisms underlying the direct involvement of miR‐506 in chemoresistance regulation have not been extensively investigated, especially in chemoresistant CRC. Thus, we sought to determine the potential mechanisms underlying the relationship between miR‐506 expression and chemoresistance‐related gene and protein expression in HCT116‐OxR CRC cells via WB and qRT‐PCR. miR‐506 expression was significantly low in HCT116‐OxR cells, while MDR1/P‐gp expression was very high. miR‐506 treatment increased the sensitivity of HCT116‐OxR cells to oxaliplatin. Moreover, HCT116‐OxR cell populations transfected with miR‐506 mimics after oxaliplatin treatment exhibited higher percentages of oxaliplatin‐induced apoptosis than cell populations transfected with miR‐Ctrl and NC vectors. We also found that miR‐506 effectively reduced MDR1/P‐gp expression, resulting in loss of the chemoresistant phenotype in HCT116‐OxR CRC cells. These findings demonstrated that miR‐506 reverses chemoresistance by down‐regulating MDR1/P‐gp expression.
The mechanism underlying the miR‐506‐mediated reversal of oxaliplatin chemoresistance is complex and remains unknown. miR‐506 may mediate MDR1 down‐regulation by regulating drug efflux, altering cell proliferation and survival or inhibiting DNA damage. In this study, we addressed the potential relationship between Wnt/β‐catenin signalling and intrinsic responsiveness to chemotherapy in HCT116‐OxR cells. As with tumorigenesis, the current evidence suggests that abnormal Wnt signalling contributes to chemoresistance in cancer via multiple mechanisms. Flahaut26 demonstrated β‐catenin transcriptional activity in chemoresistant cells via the expression of several well‐recognized Wnt/β‐catenin target genes, including cyclin‐D1 and IGF2, in neuroblastoma cells. In addition, a growing body of evidence suggests that unregulated β‐catenin activity may play a significant role in the development of resistance to traditional cytotoxic chemotherapeutic agents. Perhaps more importantly, several of these studies demonstrated reversal of resistance following inhibition or silencing of canonical Wnt signalling. Yang27 demonstrated that β‐catenin signalling inhibition leads to decreased numbers of OV6(+) cells and reverses cisplatin chemoresistance in hepatocellular carcinoma. In our study, Wnt signalling activity was up‐regulated in HCT116‐OxR cells, as was the expression and localization of β‐catenin. HCT‐116 cell resistance to oxaliplatin was reversed by miR‐506 via inhibition of β‐catenin, as shown via WB and qRT‐PCR. Some of the key downstream proteins and genes that are activated by the binding of β‐catenin to transcriptional factors of the canonical pathway include c‐MYC (MYC), cyclin D1 (CCND1), survivin (BIRC5), Axin2 (AXIN2) and matrix metalloproteinases. It was reported that Wnt/β‐catenin signalling plays an important role in the induction of MDR1/P‐gp expression.21 Shen20 revealed that inhibition of Wnt/β‐catenin signalling down‐regulates MDR1/P‐gp expression and reverses multi‐drug resistance in cholangiocarcinoma. Yamada28 suggested that MDR1 is a target gene of the TCF4/β‐catenin complex. Down‐regulation of Wnt/β‐catenin signalling repressed MDR1/P‐gp expression, inducing apoptosis in multifarious human cancer cells and validating the function of the Wnt/β‐catenin signalling pathway in CRC.28, 29 The Wnt/β‐catenin signalling pathway is altered in more than 90% of patients with CRC30 which makes it a particularly attractive therapeutic target. Hence, disruption of the Wnt/β‐catenin signalling pathway may represent an opportunity for improved CRC chemoprevention and therapy. Several miRNAs are regarded as regulators and regulate Wnt/β‐catenin signalling pathway activity.31, 32, 33, 34 However, the mechanism by which miR‐506 reverses oxaliplatin chemoresistance in CRC has not yet been elucidated. Our previous study indicated that the miR‐506‐EZH2 may suppress tumour proliferation and metastasis by inhibiting the Wnt/β‐catenin pathway. Consistent with these findings, the results of this study showed that miR‐506, Wnt/β‐catenin signalling and MDR1/P‐gp expression constitute a regulatory circuit that may be disrupted in HCT116‐OxR CRC cells. Wnt signalling is up‐regulated in HCT116‐OxR cells, as is the expression and localization of β‐catenin. We clearly demonstrated that miR‐506 overexpression resulted in MDR1 down‐regulation at the protein level, as well as down‐regulation of the downstream genes of the Wnt/β‐catenin signalling pathway. SB‐216763 is a GSK‐3β inhibitor that can activate the Wnt/β‐catenin signalling pathway and increase intracellular β‐catenin expression.35 In addition, SB‐216763, an activator of the Wnt/β‐catenin signalling pathway, repressed HCT116‐OxR‐miR‐506 cell drug susceptibility in parallel with P‐gp up‐regulation. Therefore, we concluded that miR‐506 restoration may represent a valuable therapeutic strategy because of its inhibition of MDR1/P‐gp expression via Wnt/β‐catenin signalling pathway down‐regulation in CRC.
In summary, we have demonstrated a novel miRNA‐mediated regulatory mechanism by which miR‐506 reverses MDR1/P‐gp‐mediated chemoresistance by repressing the Wnt/β‐catenin signalling pathway through β‐catenin down‐regulation. Our present study has provided strong evidence that miR‐506 up‐regulation may represent a promising chemosensitization strategy for the treatment of CRC.
Conflicts of interest
There are no conflicts of interest to disclose.
Zhou H, Lin C, Zhang Y, et al. miR‐506 enhances the sensitivity of human colorectal cancer cells to oxaliplatin by suppressing MDR1/P‐gp expression. Cell Prolif. 2017;50:e12341 10.1111/cpr.12341
Contributor Information
Yi Zhang, Email: csuzhy@aliyun.com.
Zeqiang Ren, Email: rzq0805@163.com.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- 2. Ahmed FE. Molecular markers that predict response to colon cancer therapy. Expert Rev Mol Diagn. 2005;5:353–375. [DOI] [PubMed] [Google Scholar]
- 3. Bhayani MK, Calin GA, Lai SY. Functional relevance of miRNA sequences in human disease. Mutat Res. 2012;731:14–19. [DOI] [PubMed] [Google Scholar]
- 4. Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fish JE, Santoro MM, Morton SU, et al. miR‐126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhai H, Fesler A, Ju J. MicroRNA: a third dimension in autophagy. Cell Cycle. 2013;12:246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chai H, Liu M, Tian R, Li X, Tang H. miR‐20a targets BNIP2 and contributes chemotherapeutic resistance in colorectal adenocarcinoma SW480 and SW620 cell lines. Acta Biochim Biophys Sin. 2011;43:217–225. [DOI] [PubMed] [Google Scholar]
- 9. Li PL, Zhang X, Wang LL, et al. MicroRNA‐218 is a prognostic indicator in colorectal cancer and enhances 5‐fluorouracil‐induced apoptosis by targeting BIRC5. Carcinogenesis. 2015;36:1484–1493. [DOI] [PubMed] [Google Scholar]
- 10. Li J, Chen Y, Zhao J, Kong F, Zhang Y. miR‐203 reverses chemoresistance in p53‐mutated colon cancer cells through downregulation of Akt2 expression. Cancer Lett. 2011;304:52–59. [DOI] [PubMed] [Google Scholar]
- 11. Zhong M, Bian Z, Wu Z. miR‐30a suppresses cell migration and invasion through downregulation of PIK3CD in colorectal carcinoma. Cell Physiol Biochem. 2013;31:209–218. [DOI] [PubMed] [Google Scholar]
- 12. Tong JL, Zhang CP, Nie F, et al. MicroRNA 506 regulates expression of PPAR alpha in hydroxycamptothecin‐resistant human colon cancer cells. FEBS Lett. 2011;585:3560–3568. [DOI] [PubMed] [Google Scholar]
- 13. Zhou Y, Wan G, Spizzo R, et al. miR‐203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol Oncol. 2014;8:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wan LY, Deng J, Xiang XJ, et al. miR‐320 enhances the sensitivity of human colon cancer cells to chemoradiotherapy in vitro by targeting FOXM1. Biochem Biophys Res Commun. 2015;457:125–132. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z, Ma J, Luan G, et al. MiR‐506 suppresses tumor proliferation and invasion by targeting FOXQ1 in nasopharyngeal carcinoma. PLoS ONE. 2015;10:e0122851. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Zhang Y, Lin C, Liao G, et al. MicroRNA‐506 suppresses tumor proliferation and metastasis in colon cancer by directly targeting the oncogene EZH2. Oncotarget. 2015;6:32586–32601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodel C, Martus P, Papadoupolos T, et al. Prognostic significance of tumor regression after preoperative chemoradiotherapy for rectal cancer. J Clin Oncol. 2005;23:8688–8696. [DOI] [PubMed] [Google Scholar]
- 18. Chen H, Ren C, Han C, Wang D, Chen Y, Fu D. Expression and prognostic value of miR‐486‐5p in patients with gastric adenocarcinoma. PLoS ONE. 2015;10:e0119384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu X, Wang Y, Yin PH, et al. Wnt/β‐catenin signal pathway regulates VEGF expression in human colorectal cancer cells. China Oncol. 2012;22:881–885. [Google Scholar]
- 20. Shen DY, Zhang W, Zeng X, Liu CQ. Inhibition of Wnt/beta‐catenin signaling downregulates P‐glycoprotein and reverses multi‐drug resistance of cholangiocarcinoma. Cancer Sci. 2013;104:1303–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang H, Jing X, Wu X, et al. Suppression of multidrug resistance by rosiglitazone treatment in human ovarian cancer cells through downregulation of FZD1 and MDR1 genes. Anticancer Drugs. 2015;26:706–715. [DOI] [PubMed] [Google Scholar]
- 22. Krishna R, Mayer LD. Multidrug resistance (MDR) in cancer. Mechanisms, reversal using modulators of MDR and the role of MDR modulators in influencing the pharmacokinetics of anticancer drugs. Eur J Pharm Sci. 2000;11:265–283. [DOI] [PubMed] [Google Scholar]
- 23. Montazami N, Kheir Andish M, Majidi J, et al. siRNA‐mediated silencing of MDR1 reverses the resistance to oxaliplatin in SW480/OxR colon cancer cells. Cell Mol Biol. 2015;61:98–103. [PubMed] [Google Scholar]
- 24. Liu G, Yang D, Rupaimoole R, et al. Augmentation of response to chemotherapy by microRNA‐506 through regulation of RAD51 in serous ovarian cancers. J Natl Cancer Inst. 2015;107:djv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu G, Xue F, Zhang W. miR‐506: a regulator of chemo‐sensitivity through suppression of the RAD51‐homologous recombination axis. Chin J Cancer. 2015;34:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Flahaut M, Meier R, Coulon A, et al. The Wnt receptor FZD1 mediates chemoresistance in neuroblastoma through activation of the Wnt/beta‐catenin pathway. Oncogene. 2009;28:2245–2256. [DOI] [PubMed] [Google Scholar]
- 27. Yang W, Yan HX, Chen L, et al. Wnt/beta‐catenin signaling contributes to activation of normal and tumorigenic liver progenitor cells. Cancer Res. 2008;68:4287–4295. [DOI] [PubMed] [Google Scholar]
- 28. Yamada T, Takaoka AS, Naishiro Y, et al. Transactivation of the multidrug resistance 1 gene by T‐cell factor 4/beta‐catenin complex in early colorectal carcinogenesis. Cancer Res. 2000;60:4761–4766. [PubMed] [Google Scholar]
- 29. Stein U, Fleuter C, Siegel F, et al. Impact of mutant beta‐catenin on ABCB1 expression and therapy response in colon cancer cells. Br J Cancer. 2012;106:1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shin J, Shin Y, Oh SM, et al. MiR‐29b controls fetal mouse neurogenesis by regulating ICAT‐mediated Wnt/beta‐catenin signaling. Cell Death Dis. 2014;5:e1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Strillacci A, Valerii MC, Sansone P, et al. Loss of miR‐101 expression promotes Wnt/beta‐catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol. 2013;229:379–389. [DOI] [PubMed] [Google Scholar]
- 33. Su J, Zhang A, Shi Z, et al. MicroRNA‐200a suppresses the Wnt/beta‐catenin signaling pathway by interacting with beta‐catenin. Int J Oncol. 2012;40:1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu J, Zhu X, Wu L, et al. MicroRNA‐122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/beta‐catenin pathway. Liver Int. 2012;32:752–760. [DOI] [PubMed] [Google Scholar]
- 35. Takahashi‐Yanaga F. Activator or inhibitor? GSK‐3 as a new drug target. Biochem Pharmacol. 2013;86:191–199. [DOI] [PubMed] [Google Scholar]


