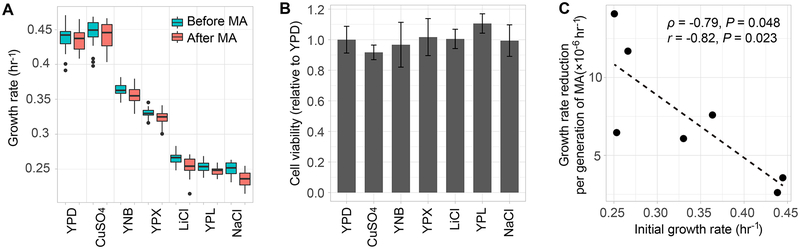
Figure 1. Growth rate and cell viability of yeast mutation accumulation (MA) lines in seven relatively benign environments.
(A) Population growth per hour before (blue) and after (red) MA in each environment. The data are presented as a bar plot for each bin. The lower and upper edges of a box represent the first (qu1) and third quartiles (qu3), respectively, while the horizontal line inside the box indicates the median (md). The whiskers extend to the most extreme values inside inner fences, md±1.5(qu3- qu1), and the dots represent values outside the inner fences (outliers).
(B) Cell viability in each environment, relative to that in YPD. Error bars represent the standard deviation. In panels (A) and (B), the six non-YPD environments are ordered from high to low growth rates on the X-axis.
(C) Negative correlation between the speed of growth rate decline in MA and the initial growth rate across environments. The speed of growth rate decline in MA is (growth rate before MA – growth rate after MA)/number of cell divisions. Because of the use of the growth rate before MA in both variables of the X- and Y-axes, the actual correlation could be even more negative than what is observed here. Spearman’s ρ and Pearson’s r are shown in the figure with corresponding P values.
See also Table S1.