Abstract
The troponin complex plays a central role in regulating the contraction and relaxation of striated muscles. Among the three protein subunits of troponin, the calcium receptor subunit, TnC, belongs to the calmodulin family of calcium signaling proteins whereas the inhibitory subunit, TnI, and tropomyosin-binding/thin filament-anchoring subunit, TnT, are striated muscle-specific regulatory proteins. TnI and TnT emerged early in bilateral symmetric invertebrate animals and have co-evolved during the 500–700 million years of muscle evolution. To understand the divergence as well as conservation of the structures of TnI and TnT in invertebrate and vertebrate organisms adds novel insights into the structure-function relationship of troponin and the muscle type isoforms of TnI and TnT. Based on the significant growth of genomic database of multiple species in the past decade, this focused review studied the primary structure features of invertebrate troponin subunits in comparisons with the vertebrate counterparts. The evolutionary data demonstrate valuable information for a better understanding of the thin filament regulation of striated muscle contractility in health and diseases.
Keywords: Troponin isoforms, invertebrate muscle, molecular evolution, myofilament, TnI, TnT
Muscle contraction is a vital function of animals. Striated muscles have a highly organized contractile machinery consisting of sarcomeres as the basic contractile units formed with overlapping myosin thick filaments and actin thin filaments. Straited muscle contraction is powered by actin-activated myosin ATPase under the regulation of intracellular Ca2+ via the thin filament-associated troponin complex.
The troponin complex contains three protein subunits: The calcium receptor subunit, TnC, the inhibitory subunit, TnI, and the tropomyosin-binding/thin filament-anchoring subunit, TnT. To study the molecular evolution of troponin helps to understand the structure-function relationship of troponin and muscle contraction. This focused review discusses the divergence as well as conservation of troponin protein in invertebrate and vertebrate species for insights into the evolution and contractile feature of different types of striated muscle.
1. Striated Muscles and Troponin Regulation of Contraction
a). Three types of vertebrate muscle
Three distinct types of muscle cells exist in vertebrates: Skeletal, cardiac and smooth. Skeletal muscle associates with the skeleton to power voluntary movement. Cardiac muscle in the walls of the heart chambers pumps blood through the circulatory system. Skeletal and cardiac muscles are striated muscles. While smooth muscle also contains myosin thick filament and actin thin filament based contractile machinery, the myofilaments in smooth muscle cells are not organized into sarcomeres, corresponding to its non-striated appearance (1).
b). The types of invertebrate muscles
The muscle of invertebrate animals can also be divided into striated and smooth types based on the presence or absence of Z-discs as a defining structure of sarcomeres. Sarcomere structures of vertebrate and invertebrate striated muscles are similar and analogous (2). The invertebrate striated muscles are further sub-grouped into transverse and oblique muscles according to the different patterns of striation. Invertebrate transverse striated muscle is very similar to vertebrate striated muscle with thick and thin filaments organized into sarcomeres (3). As that in vertebrate striated muscles, the actin thin filaments in invertebrate striated muscle sarcomeres contain troponin complex that functions in the Ca2+-regulation of contraction and relaxation.
c). The classic model of calcium-troponin regulation in vertebrate striated muscle
The contraction of cardiac and skeletal muscles in vertebrate animals plays vital physiological functions. The contraction and relaxation of striated muscle cells are regulated by Ca2+ via the actin filament-associated troponin-tropomyosin system (4, 5). Contraction is initiated by a rising of cytosolic Ca2+ following depolarization of plasma membrane and Ca2+ release from the sarcoplasmic reticulum. Ca2+-binding to TnC induces conformational changes that are transduced to TnI and TnT to release the tropomyosin block of myosin-binding sites on the actin thin filament (6, 7). Binding of myosin heads extending from the thick filament to actin thin filament form cross bridges and activates myosin ATPase to generate power strokes that slide the thin filaments towards the center of the sarcomere and result in contraction (5, 6).
2. Conserved Contractile Machinery of Striated Muscles and Divergence between Vertebrates and Invertebrates
a). The Sarcomeres
In vertebrates, the contractile machinery of striated muscles is the myofibril consisting of tandem repeats of sarcomeres that are the basic contractile units composed of overlapping myosin thick filaments and actin thin filaments (4, 5). This organization is the basis for the thick and thin filament sliding model of striated muscle contraction and is conserved between vertebrates and invertebrates although myofilament arrangement differs in some invertebrate species (e.g., sarcomeres of C. elegans body wall muscle are obliquely aligned (8)).
The myofilament protein contents in vertebrate and invertebrate striated muscle sarcomeres are similar. In vertebrates, the thick filament is composed of myosin organized into bi-polar polymers with their globular heads toward the two ends of the filaments. The actin thin filament contains tropomyosin and troponin. Similarly, tropomyosin and troponin are associated with the actin thin filaments in invertebrate striated muscle sarcomeres.
There is also a notable divergence between vertebrate and invertebrate striated muscle sarcomeres. In contrast to that of vertebrates, the invertebrate muscle thick filaments have variable length and width. The thick myofilaments of some invertebrate muscles contain a central core of paramyosin, which is absent in vertebrate muscles (2).
The actin thin filaments-associated Ca2+-troponin-tropomyosin system regulates the actin-myosin interaction and ATPase cross bridge activation in C. elegans body wall muscle (8) and insect indirect flight muscle (9). In addition to the troponin-based thin filament regulation, mollusc catch muscle contraction is also regulated through the thick filaments (10), in which Са2+ regulates phosphorylation of myosin and activates ATPase, resembling that in vertebrate smooth muscles (11).
b). Early emergence of troponin during animal evolution
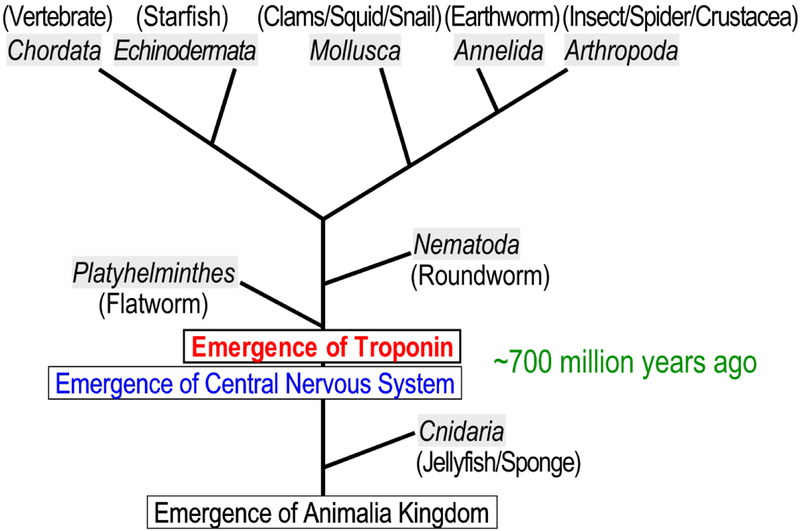
Troponin plays a central role in the regulation of striated muscle contraction and relaxation (7, 12). Genes that encode the subunits of troponin are found in all invertebrate animals except for Cnidaria (jellyfish and sponges), indicating an early emergence approximately 700 million years ago during the evolution of animals (Fig. 1). This timeline corresponds to the emergence of bilaterians in animals (the phyla of Platyhelminthes and Nematoda), concurrent with the emergence of central nervous system (13, 14). This notion suggests a critical role of the troponin regulation of muscle contraction and relaxation in coordinated movement of animals.
Fig. 1. Early emergence of troponin during the evolution of animals.
Based on available sequence data of animal genomes, this unscaled illustration of phylogenic lineage of the phyla of animal kingdom illustrates the emergence of troponin ~700 million years ago prior to the emergence of Platyhelminthes and Nematoda.
3. Evolutionary Lineage of Troponin Subunits
Among the three subunits of vertebrate troponin, TnC has two isoforms expressed in slow skeletal/cardiac muscles (TnnC1) and fast skeletal muscle (TnnC2), respectively (15). While TnC belongs to the calmodulin family of E-F hand calcium binding proteins, TnI and TnT are co-evolved striated muscle-specific proteins (16, 17). Vertebrate TnI and TnT have both diverged into three muscle type-specific isoforms: Tnni1 and Tnnt1 in slow twitch skeletal muscle, Tnni2 and Tnnt3 in fast twitch skeletal muscle, and Tnni3 and Tnnt2 in cardiac muscle. Consistent with their co-evolutionary relationship, genes encoding TnI and TnT in vertebrate genomes are closely linked in three physical pairs (Tnni1-Tnnt2, Tnni2-Tnnt3 and Tnni3-Tnnt1), indicating the origin of TnI and TnT from the duplication of a single ancestral gene (17).
Like that in vertebrate striated muscles, the troponin complex is associated with the sarcomeric thin filament in invertebrate striated muscles to play a central role in the Ca2+ regulation of contraction and relaxation. The three subunits of invertebrate troponin are encoded by three separate genes: TpnC for TnC, TpnI for TnI and TpnT for TnT. While two TnC isoform genes are present in vertebrates, six copies of TpnC gene are found in insects (18). In contrast to the diverged muscle type-specific TnI and TnT isoform genes in vertebrates, only one TnI gene and one TnT gene are found in invertebrates (19).
Although significantly diverged in overall primary structures, invertebrate and vertebrate troponin subunits have conserved core structures and similar structure-function relationships. The posttranscriptional regulation of invertebrate and vertebrate troponin subunits also employs conserved mechanisms. As discussed below, the generation of TnT variants via alternative RNA splicing is found in both vertebrate and invertebrate muscles (7, 19).
4. Invertebrate TnC
TnC is the Ca2+ receptor of troponin. The overall primary structures of invertebrate and vertebrate TnC are diverged (Fig. 2A). A homolog of calmodulin, invertebrate and vertebrate TnC have four conserved EF-hand metal ion binding sites (Fig. 2B) (20). Ca2+ binding to TnC results in conformational changes, which is relayed to TnI, TnT and the tropomyosin-action thin filament to release the inhibition of actin-myosin interaction and induce muscle contraction (5). Different from the Ca2+ regulation via the N domain of TnC in vertebrates, the activation of mollusca muscle contraction is uniquely via Ca2+ binding to the C domain of TnC (21). For the conserved role of TnC as a calmodulin-like calcium receptor other than a striated muscle-specific regulatory protein, its evolution and structure-function relationship in invertebrate muscles is not discussed in detail in this focused review.
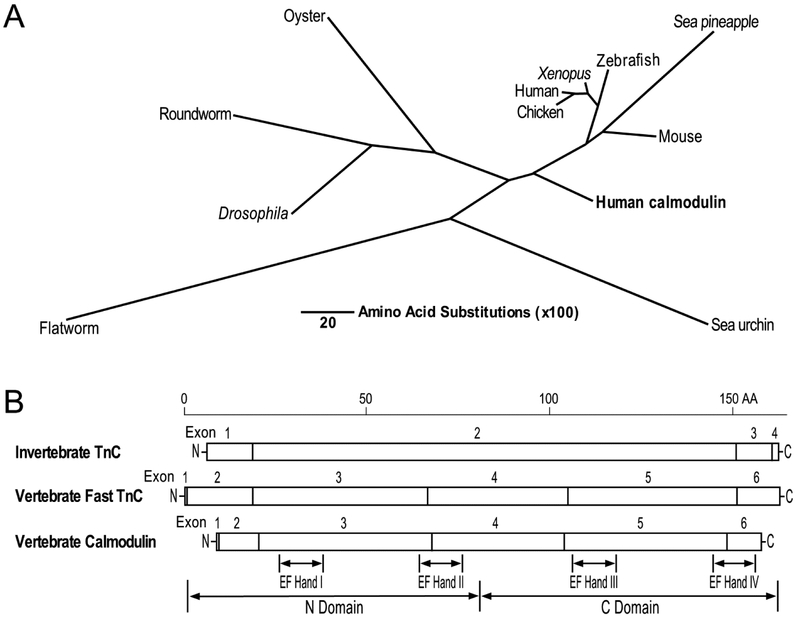
Fig. 2. Evolution and structure features of TnC in vertebrates and invertebrates.
(A) The phylogenetic tree of TnC constructed by aligning amino acid sequences from representative species of animal phyla using the Clustal V method of DNAStar MegAlign software shows the divergence of TnC between invertebrates and vertebrates as well as the homology to calmodulin. The length of each pair of branches represents the distance between sequence pairs, while the scale bar indicating the distance corresponding to 20 amino acid substitutions per 100 residues. The sequence accession numbers are: Sea urchin TnC, AAA30007.1; flatworm TnC, XP_012797142.1; Drosophila TnC, NP_523619.2; roundworm TnC, BAA82523.1; Oyster TnC, BBD82024.1; chicken TnC, AAA49097.1; human TnC, AAA91854.1; Xenopus TnC, NP_001079408.1; zebrafish TnC, AAH64284.1; sea pineapple TnC, BAA13630.1; mouse TnC, NP_033419; human calmodulin, CAA36839. (B) The linear maps of protein primary structures illustrate the similarity between invertebrate and vertebrate TnC and to calmodulin with the four EF hand metal binding sites in the N and C domains indicated.
5. Invertebrate TnI
TnI is the ATPase inhibitory subunit of troponin (22). In contrast to the three muscle type-specific isoforms of TnI in vertebrates, only one TnI gene exists in invertebrates. Evolutionary lineage studies have indicated that the fast skeletal muscle TnI represents the ancestral form of vertebrate TnI (17). Therefore, we may use vertebrate fast TnI sequences to compare with that of invertebrate TnI to investigate their evolutionary and structure-function relationships.
The phylogenetic tree in Fig. 3A constructed from protein sequences demonstrates the significant divergence between vertebrate and invertebrate TnI genes. On the other hand, the protein primary structure maps in Fig. 3B and the DotPlot sequence alignment in Fig. 3C illustrate conserved core structures in vertebrate and invertebrate TnI. An N-terminal extension is present in vertebrate cardiac TnI and invertebrate TnI. The N-terminal extension of vertebrate cardiac TnI has been extensively studied for its role in regulating cardiac muscle contractility and heart function via b-adrenergic stimulated PKA phosphorylation sites (7, 22).
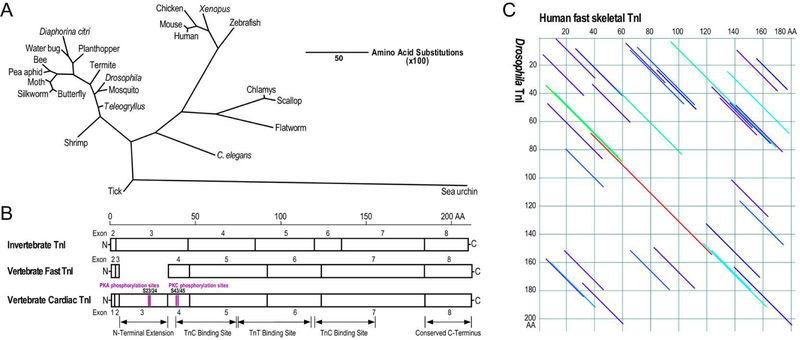
Fig. 3. Evolution and structure features of TnI in vertebrates and invertebrates.
(A) The phylogenetic tree of TnI constructed by aligning amino acid sequences from representative species of animal phyla using the Clustal V method of DNAStar MegAlign software demonstrates significant evolutionary divergence while vertebrates and insects being conserved clusters. The length of each pair of branches represents the distance between sequence pairs, while the scale bar indicating the distance corresponding to 50 amino acid substitutions per 100 residues. The sequence accession numbers are: Sea urchin TnI, XP_011664558.1; tick TnI, BAB55451.1; shrimp TnI, ACV40756.1; butterfly TnI, NP_001299300.1; silkworm TnI, NP_001037295.1; moth TnI, KOB71297.1; pea aphid TnI, NP_001313576.1; bee TnI, NP_001035346.1; water bug TnI, CAF18234.1; Diaphorina citri TnI, ABG81999.1; planthopper TnI, ACN79503.1; termite TnI, KDQ88314.1; Drosophila TnI, CAA42020.1; mosquito TnI, XP_001864736.1; teleogryllus TnI, AVI126882.1; human fast TnI, AAH32148.1; mouse TnI, NP_033431.1; chicken fast TnI, AAA61952.1; Xenopus fast TnI, AAH84508.1; zebrafish fast TnI, AAI62242.1; chlamys TnI, BAE43658.1; scallop TnI, BAE43658.1; flatworm TnI CAX73588.1; C. elegans TnI, NP_509906.1. B) The linear maps of protein primary structure demonstrate the conserved core structures of vertebrate and invertebrate TnI containing TnC and TnT binding sites. A regulatory N-terminal extension is present in vertebrate cardiac TnI. Invertebrate TnI also has an N-terminal extension although its function has not been extensively studied. C) Paired DotPlot alignment of amino acid sequences of human fast skeletal muscle TnI (accession # AAH32148.1) and Drosophila TnI (accession # CAA42020.1) demonstrates the conserved core structures. The diagonal lines indicate regions of the two sequences which meet the threshold for similarity specified in the parameters set for the analysis, of which dark blue indicates weak similarity with progressively stronger similarities shown in light blue, green, yellow, orange and red. The long red diagonal line indicates the region containing conserved binding sites for TnC and TnT (B).
The function of the N-terminal extension of invertebrate TnI remains to be investigated. A study reported that it is required in C. elegans striated muscle for coordinated worm locomotion (13). Although it is unclear whether the N-terminal extensions of invertebrate TnI and vertebrate cardiac TnI are from a conserved ancestral structure or diverged structures, it may act as a site to modulate the overall conformation and function of TnI as shown vertebrate cardiac TnI (23).
6. Invertebrate TnT
TnT is the tropomyosin-binding and thin filament anchoring subunit of troponin (24). Co-evolved with TnI isoform genes, fast skeletal muscle TnT represents the ancestral form of vertebrate TnT isoforms (17), which is compared with invertebrate TnT for their evolutionary and structure-function relationships.
A phylogenetic tree constructed from protein sequences of representative vertebrate and invertebrate TnTs is shown in Fig. 4A. The evolutionary lineage demonstrates significant divergence among animal phyla. The phylogenetic analysis also showed notable conservation of TnT primary structure within the phylum of vertebrates and the class of insects. These evolutionary divergence and conservation implicate a role of TnT function in adaptation to the functional features of different animal muscles.
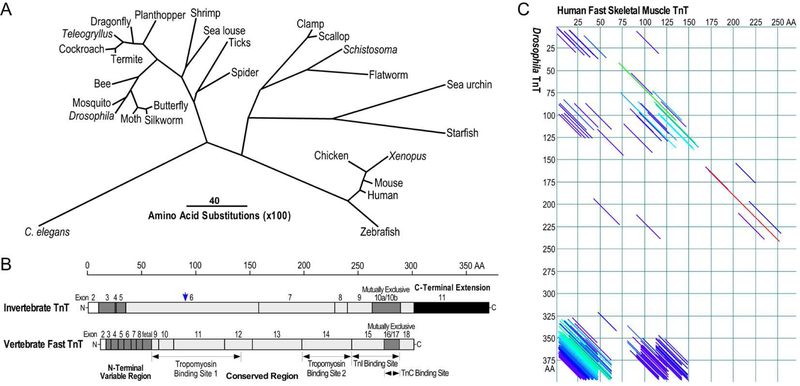
Fig. 4. Evolution and structure features of TnT in vertebrates and invertebrates.
(A) The phylogenetic tree of TnT constructed by aligning amino acid sequences from representative species in animal phyla using the Clustal V method of DNAStar MegAlign software shows significant evolutionary divergence. The length of each pair of branches represents the distance between sequence pairs, while the scale bar indicating the distance corresponding to 40 amino acid substitutions per 100 residues. Similar to the pattern of TnI shown in Fig. 3A, vertebrate TnT and insect TnT are conserved in clusters. The sequence accession numbers are: C. elegans TnT, NP_001024704.1; butterfly TnT, BAG30738.1; silkworm TnT, ABD36267.1; moth TnT, ADO33067.1; Drosophila TnT, NP_001162742.1; mosquito TnT, XP_001851541.1; bee TnT, NP_001035348.1; termite TnT, AGM32088.1; cockroaches TnT, AAD33603.1; Teleogryllus TnT, AVI26881.1; dragonfly TnT, AAD33604.1; planthopper TnT, AGI96988.1; shrimp TnT, AQV08184.2; sea louse TnT, ACO12887.1; tick TnT, AAY42205.1; spider TnT, EU247211.1; clamp TnT, BAA13610.1; scallop TnT, BAA20456.1; Schistosoma TnT, XP_018646291.1; flatworm TnT, XP_024354240.1; sea urchin TnT, XP_011671209.1; ; starfish TnT, XP_022088338.1; chicken TnT, NP_990253.1; Xenopus TnT, NP_989143.1; mouse TnT, NP_001347086.1; human TnT, NP_001354775.1; fish TnT, AAF78472.1. B) The linear maps of protein primary structure outline the divergence as well as conservation between vertebrate and invertebrate TnT with their middle and C-terminal conserved regions (highlighted in gray) containing binding sites for tropomyosin, TnI and TnC, an N-terminal variable region, a C-terminal mutually spliced segment, and a C-terminal extension unique to invertebrate TnT, especially insect TnT. The alternatively spliced exons are shown as hatched boxes whereas the C-terminal extension of invertebrate TnT is highlighted in solid black. The alternative translational initiation site that generates N-terminal truncated TnT in Drosophila is indicated with a blue arrow. C) Paired DotPlot alignment of amino acid sequences of human fast skeletal muscle TnT (accession # NP_001354775.1) and Drosophila TnT (accession # NP_001162742.1) demonstrates the middle and C-terminal conserved regions. The diagonal lines indicate regions of the two sequences which meet the threshold for similarity specified in the parameters set for the analysis, of which dark blue indicates weak similarity with progressively stronger similarities shown in light blue, green, yellow, orange and red. Although non-homologous structures, the N-terminal variable region of vertebrate TnT and the C-terminal extension of insect TnT showed a strong similarity based on their unique high contents of Glu residues.
The protein primary structure maps in Fig. 4B show that the middle and C-terminal (except the invertebrate C-terminal extension) regions form the TnT core structures and are conserved between vertebrates and invertebrates. The N-terminal domain is a variable region in both vertebrate and invertebrate TnT. Extensive studies of vertebrate TnT have demonstrated that the N-terminal variable region is highly diverged among the three isoform genes and across species (7, 24). Structure of the N-terminal variable region of all three vertebrate TnT isoforms is also regulated via alternative RNA splicing in different muscle types and during heart and skeletal muscle development (7, 24, 25). Alternative splicing of N-terminal coding exons also generates protein variations in Drosophila TnT (26). 5’ truncated variants of Drosophila TpnT mRNA have been found to encode N-terminal truncated protein, apparently using an alternative translational initiation site in exon 6 (Fig. 4B). Studies of vertebrate TnT demonstrated that the N-terminal variable region is a regulatory structure that modulates the conformation and function of TnT to tune muscle contractility (24, 27).
A pair of mutually exclusive exons (exons 16 and 17) are present in vertebrate fast skeletal muscle TnT but not in cardiac or slow TnT (24, 25, 28). A similar pair of mutually exclusive C-terminal coding exons (exons 10a and 10b) are found in the Drosophila TpnT gene (19) (Fig. 4B). This conserved feature that emerged early before the divergence of vertebrates and invertebrates supports the notion that fast skeletal muscle TnT represents the ancestral form of vertebrate TnT while the exon 16 encoded variant was lost later during the evolution of cardiac and slow TnT genes (17). The segment encoded by these mutually exclusive exons is in the conserved core structure of TnT and the functional significance of the alternative splicing regulation is worth investigating.
Invertebrate TnT has a unique C-terminal extension (Fig. 4B) that contains up to more than 50 glutamic acids in insect TnT (29). Its function is unknown. Intriguingly, the N-terminal variable region of vertebrate TnT also has high contents of Glu residues (7). This feature is most clear in the alternative splice-forms of fast skeletal muscle TnT expressed in avian pectoral muscles (30, 31). This sequence similarity between the C-terminal extension of invertebrate TnT and the N-terminal variable region of vertebrate TnT is represented in the DotPlot sequence alignment in Fig. 4C. The glutamic acid rich segments may have resulted from similar evolutionary selections and their potentially analogous functions are also worth investigating.
7. Summary
With the rapidly growing genomic database of multiple species in the past decade, looking into the evolution and structure of invertebrate troponin in comparison with the vertebrate counterparts can give us insightful information for the structure-function relationship of troponin and a better understanding of the regulation of muscle contraction. Data summarized in this focused review demonstrate that the N-terminal extension is a regulatory site of TnI conserved in vertebrates and invertebrates. Consistent with their common origin from the duplication of a single ancestor gene (17), the N-terminal variable region of TnT is also a conformational modulator conserved in vertebrates and invertebrates.
The mutually exclusively spliced exons 16/17 in vertebrate fast TnT and exons 10a/10b in invertebrate TnT represent another conserved trait that emerged before the divergence of vertebrates and invertebrates. The later loss of exon 16 in vertebrate cardiac TnT and slow TnT genes (7, 24, 25) adds an evidence for the elimination of exons during the evolution of TnT isoform genes as shown by the fewer exons in vertebrate slow skeletal muscle TnT gene than that in fast and cardiac TnT genes (17). The preservation of exon 16 in vertebrate fast skeletal TnT may indicate a value specific to fast twitch contractions. To examine the expression of exon 10a or 10b TnT in different invertebrate muscle types may help to understand the functional significance.
Previous studies of vertebrate TnT have demonstrated that the primary structure of TnT is more diverged between the three muscle type isoforms than that across species (25), supporting functional selection vs. genetic drifting. Therefore, the Glu-rich C-terminal extension in insect TnT and the N-terminal segment in avian fast TnT (7, 31) may relate to a similar role in muscle contractility during flight. The insect indirect flight muscle functions based on stretch activation at an intermediate level of cytosolic Ca2+, allowing alternating stretch of opposing muscles to produce rapid oscillatory wing beats in Drosophila (32, 33) and in larger insects with lower wing beat frequencies such as the giant water bug, Lethocerus (33). A previous study showed that the N-terminal Glu-rich segment of avian pectoral muscle TnT has a Ca2+ binding capacity (34). The potential contribution of the Glu-rich segments of insect and avian TnT to myocyte Ca2+ handling and the stretch activation mechanism presents a novel hypothesis for future research.
Troponin plays a central role in striated muscle contraction and relaxation. Structural divergence and conservation of TnI and TnT in invertebrates and vertebrates reflect adaptations in the thin filament regulation. The information discussed here helps to understand physiological modifications and pathological mutations of troponin and guides the development of targeted treatment for human muscle and heart diseases.
Acknowledgement
This work is supported in part by grants from the National Institutes of Health (HL-127691 and 138007 to JPJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sanger JW, Wang J, Fan Y, White J, Mi-Mi L, Dube DK, Sanger JM, Pruyne D. Assembly and maintenance of myofibrils in striated muscle. Handbook of Experimental Pharmacology. 2017; 235:39–75. [DOI] [PubMed] [Google Scholar]
- 2.Perkins AD, Tanentzapf G. An ongoing role for structural sarcomeric components in maintaining Drosophila melanogaster muscle function and structure. PLoS One. 2014; 9:e99362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paniagua R, Royuela M, García-Anchuelo RM, Fraile B. Ultrastructure of invertebrate muscle cell types. Histol Histopathol. 1996; 11:181–201. [PubMed] [Google Scholar]
- 4.Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996; 58:447–481. [DOI] [PubMed] [Google Scholar]
- 5.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000; 80:853–924. [DOI] [PubMed] [Google Scholar]
- 6.Rassier DE. Sarcomere mechanics in striated muscles: From molecules to sarcomeres to cells. Am J Physiol Cell Physiol. 2017; 313:C134–45. [DOI] [PubMed] [Google Scholar]
- 7.Jin JP, Zhang Z, Bautista JA. Isoform Diversity, Regulation, and Functional Adaptation of Troponin and Calponin, Eukaryotik Gene Expression, 2008; 18:93–124. [DOI] [PubMed] [Google Scholar]
- 8.Ono Shoichiro. Regulation of structure and function of sarcomeric actin filaments in striated muscle of the nematode Caenorhabditis elegans. Anatomical record (Hoboken, NJ) 2014; 297:1548–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullard B, Leonard K, Larkins A, Butcher G, Karlik C, Fyrberg E. Troponin of asynchronous flight muscle. J. Mol Biol 1988; 204:621–637. [DOI] [PubMed] [Google Scholar]
- 10.Shelud’ko NS, Matusovskaya GG, Permyakova TV, Matusovsky OS. Twitchin a thick-filament protein from molluscan catch muscle, interacts with F-actin in a phosphorylation-dependent way. Arch. Biochem. Biophys, 2004; 432:269–277. [DOI] [PubMed] [Google Scholar]
- 11.Kim HR, Appel S, Vetterkind S, Gangopadhyay SS, Morgan KG. Smooth muscle signaling pathways in health and disease. J Cell Mol Med. 2008; 2:2165–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng JJ, Jin JP. Isoform, Splice-form, and Posttranslational Regulations of Troponin Subunits in Cardiac Development and Adaptation: A Focused Review. Frontiers in Striated Muscle Physiology 2014; 5:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes DE, Hwang H, Ono K, Lu H, Ono S. Molecular evolution of troponin I and a role of its N-terminal extension in nematode locomotion. Cytoskeleton (Hoboken, NJ). 2016; 73:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kagawa H, Takaya T, Ruksana R, Anokye-Danso F, Amin MZ, Terami H. C. elegans model for studying tropomyosin and troponin regulations of muscle contraction and animal behavior. Adv Exp Med Biol. 2007; 592:153–61. [DOI] [PubMed] [Google Scholar]
- 15.Gillis TE, Marshall CR, Tibbits GF. Functional and evolutionary relationships of troponin C. Physiol. Genomics 2007; 32:16–27. [DOI] [PubMed] [Google Scholar]
- 16.Herranz R, Mateos J, Mas JA, Garcia-Zaragoza E, Cervera M, Marco R. The coevolution of insect muscle TpnT and TpnI gene isoforms. Mol Biol Evol. 2005; 22:2231–2242. [DOI] [PubMed] [Google Scholar]
- 17.Chong SM, Jin JP. To investigate protein evolution by detecting suppressed epitope structures. J Mol Evol. 2009; 68:448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herranz R, Mateos J, Marco R. Diversification and independent evolution of TpnC genes in insects. J. Mol. Evol 2005; 60:31–44. [DOI] [PubMed] [Google Scholar]
- 19.Nongthomba U, Ansari M, Thimmaiya D, Stark M, Sparrow J Aberrant splicing of an alternative exon in the Drosophila troponin-T gene affects flight muscle development. Genetics. 2007; 177:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabarek Z Structural basis for diversity of the EF-hand calcium-binding proteins. Journal of molecular biology. 2006; 359:509–525. [DOI] [PubMed] [Google Scholar]
- 21.Doi T, Satoh A, Tanaka H, Inoue A, Yumoto F, Tanokura M, Ohtsuki I, Nishita K, Ojima T. Functional importance of Ca2+-deficient N-terminal lobe of molluscan troponin C in troponin regulation. Arch. Biochem. Biophys 2005; 436:83–90. [DOI] [PubMed] [Google Scholar]
- 22.Sheng JJ, Jin JP. TNNI1, TNNI2 and TNNI3: Evolution, regulation, and protein structure-function relationships. Gene. 2015; 576:385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akhter S, Bueltmann K Jr, Huang X, Jin JP. Restrictive cardiomyopathy mutations demonstrate functions of the C-terminal end-segment of troponin I. Arch Biochem Biophys. 2014; 552–553:3–10. [DOI] [PubMed] [Google Scholar]
- 24.Wei B, Jin, JP. Troponin T: Isoform Genes, Regulation, and Protein Structure-Function Relationships. Gene. 2016; 582:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin JP, Chen A, Huang QQ. Three alternatively spliced mouse slow skeletal muscle troponin T isoforms: conserved primary structure and regulated expression during postnatal development. Gene. 1998; 214(1–2):121–129. [DOI] [PubMed] [Google Scholar]
- 26.Domingo A, GonzáLez-Jurado J, Maroto M, Diáz C, Vinós J, Carrasco C, Cervera M, Marco R. Troponin-T is a calcium-binding protein in insect muscle: In vivo phosphorylation, muscle-specific isoforms and developmental profile in Drosophila melanogaster. J Muscle Res Cell Motil. 1998; 19:393–403. [DOI] [PubMed] [Google Scholar]
- 27.Feng HZ, Biesiadecki BJ, Yu ZB, Hossain MM, Jin JP. Restricted N-terminal truncation of cardiac troponin T: A novel mechanism for functional adaptation to energetic crisis. J Physiol (London). 2008; 586:3537–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Jin JP. Primary structure and developmental acidic to basic transition of 13 alternatively spliced mouse fast skeletal muscle troponin T isoforms. Gene. 1997; 193:105–114. [DOI] [PubMed] [Google Scholar]
- 29.Fyrberg E, Fyrberg CC, Beall C, Saville DL. Drosophila melanogaster troponin-T mutations engender three distinct syndromes of myofibrillar abnormalities. J Mol Biol. 1990; 216:657–675. [DOI] [PubMed] [Google Scholar]
- 30.Ogut O, Granzier H, Jin JP. Acidic and basic troponin T isoforms in mature fast-twitch skeletal muscle and effect on contractility. Am J Phys. 1999; 276:1162–1170. [DOI] [PubMed] [Google Scholar]
- 31.Jin JP, Samanez RA. Evolution of a metal-binding cluster in the NH(2)-terminal variable region of avian fast skeletal muscle troponin T: functional divergence on the basis of tolerance to structural drifting. J Mol Evol. 2001; 52:103–116. [DOI] [PubMed] [Google Scholar]
- 32.Singh SH, Kumar P, Ramachandra NB, and Nongthomba U Roles of the troponin isoforms during indirect flight muscle development in Drosophila. J. Genet 2014; 93: 379–388. [DOI] [PubMed] [Google Scholar]
- 33.Agianian B, Krzic U, Qiu F, Linke WA, Leonard K, Bullard B. A troponin switch that regulates muscle contraction by stretch instead of calcium. EMBO J. 2004; 23:772–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Jin, JP, and Root DD. Binding of Calcium Ions to An Avian Flight Muscle Troponin T. Biochemistry. 2004; 43:2645–55. [DOI] [PubMed] [Google Scholar]