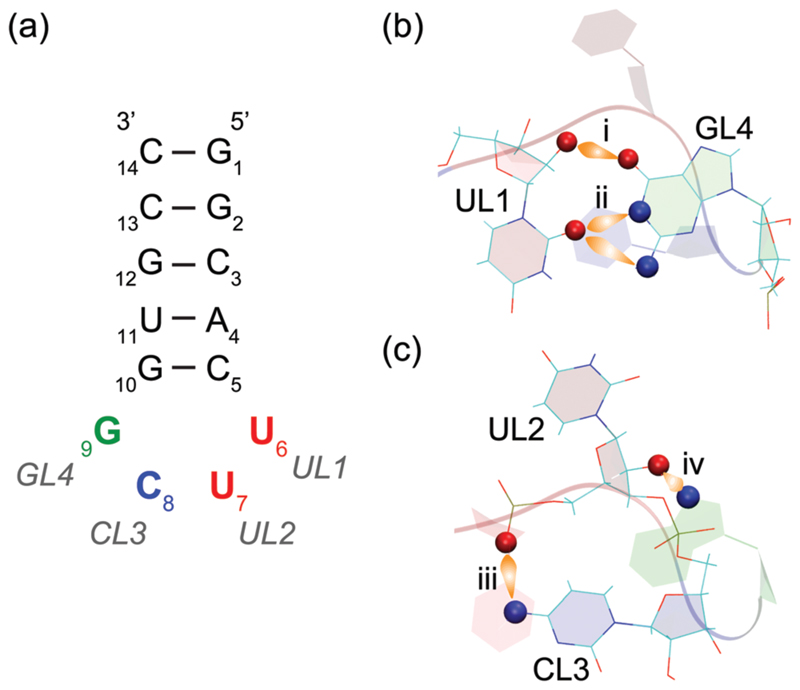
Fig. 1.
Schematic representation of the 14-nucleotide RNA hairpin containing the UUCG tetraloop that we studied in this work. (a) The nucleobases of the tetraloop are referred to as UL1, UL2, CL3 and GL4. (b and c) Signature hydrogen bond interactions and residue orientations in the UUCG tetraloop. (b) UL1 and GL4 are base-paired via (i) UL1 (O2′)…GL4(O6) and (ii) UL1(O2)…GL4(N1 and/or N2). (c) Additionally, base-backbone interactions appear via (iii) CL3(N4)…UL2(phosphate) and (iv) GL4(N7)…UL2(O2′). This network of hydrogen bonds between the tetraloop residues confers high thermodynamic stability to this structural motif. The GL4 base is in the syn conformation (i.e. with −90° < χ < 90°, where the dihedral angle χ defines the rotation around the ribose-nucleobase glycosidic bond). CL3 stacks under UL1, while UL2 remains unpaired, non-stacked and exposed to the solvent. Both UL2 and CL3 are in a C2′ -endo conformation of the sugar pucker that facilitates the bending of the RNA backbone at the tetraloop.