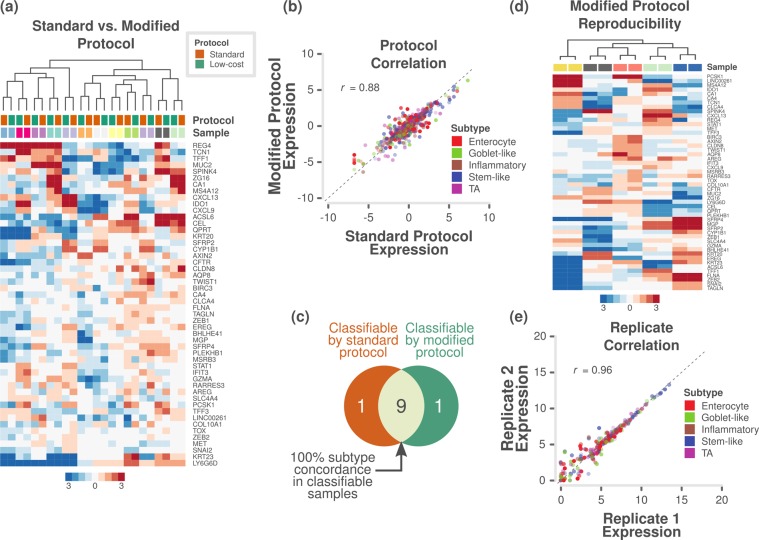
Figure 2.
Assessment of protocols and reproducibility of reduced gene subtype-based nCounter assay in FFPE samples. (a) Heatmap of expression levels of the selected 48 subtype-specific genes (and 2 additional genes) for 12 patient samples from the RETRO-C cohort as measured on a custom nCounter panel using both standard and modified protocols (24 samples – 12 each from two protocols; median centred within protocols before clustering). (b) A scatter plot of gene expression measurements for 48 genes in 12 samples between the standard and modified protocols (median centred within protocols before correlation). Each point is coloured by the gene’s weight and subtype (PAM score) in the CRCA-786 centroids. Correlation co-efficient (Pearson’s r) value is shown. (c) Venn diagram indicating the number of samples that were classifiable by the standard and modified protocols, and the concordance between classifiable samples. (d) Heatmap of expression levels of the selected 48 subtype-specific genes (and 2 additional genes) from 5 technical duplicates assayed using modified protocol with a maximum interval of 13 weeks to show the reproducibility of the assay. (e) A scatter plot of gene expression measurements for 48 genes in 5 samples between technical duplicates. Each point is coloured by the gene’s weight and subtype (PAM score) in the CRCA-786 centroids. Correlation co-efficient (Pearson’s r) value is shown.