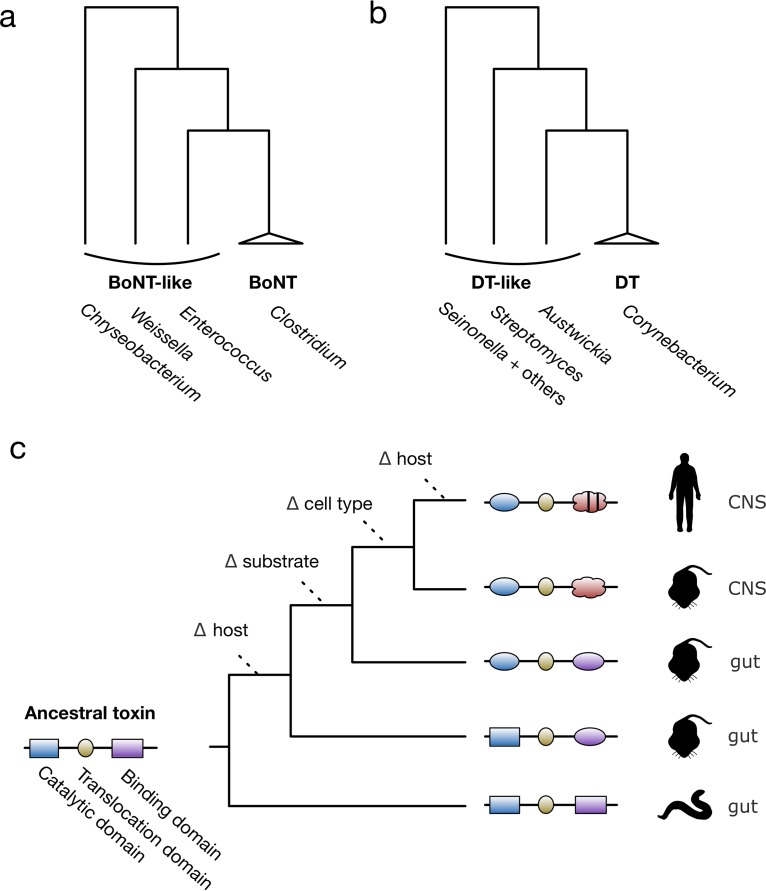
FIG 1.
Bioinformatically identified toxins provide insights into the evolutionary origins of major toxin families. (a and b) General overviews of the phylogenies of botulinum neurotoxin (BoNT) and diphtheria toxin (DT), including recently discovered homologs. (c) General model depicting the hypothetical evolution of an ancestral toxin that diversifies over time in terms of host, cell type, and substrate specificity. Changes in the catalytic domain (blue) are associated with alterations in substrate specificity, while changes in the binding domain (purple or red) are associated with alterations in host specificity and cell/tissue type (e.g., from the gut to the central nervous system [CNS]). This model is one potential explanation for observed sequence patterns in these toxin families.