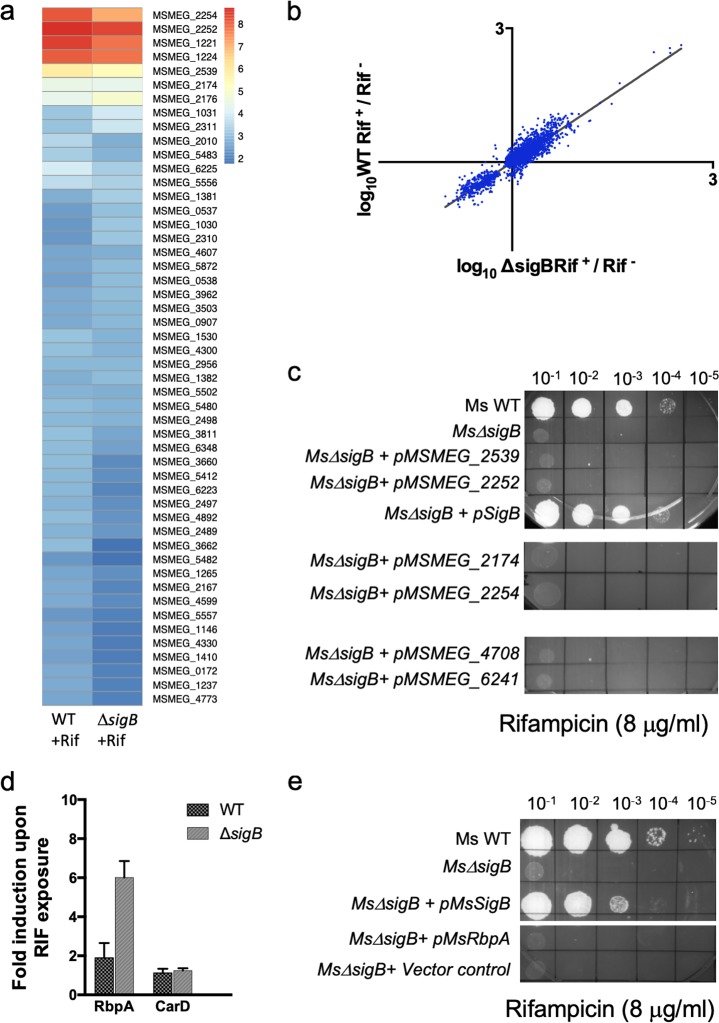
FIG 3.
Transcriptomic changes accompanying RIF exposure in the wild-type and ΔsigB mutant M. smegmatis strains. (a) Wild-type M. smegmatis and the ΔsigB mutant were exposed to 4 μg/ml of RIF for 20 min and analyzed using RNA-seq. Unexposed samples of both strains were used as controls. Two biological replicates of each sample were used. Genes induced >4-fold with a q value of <0.001 were analyzed further, and the 50 most induced genes are represented as a heat map. (Left) WT; (right) ΔsigB mutant. (b) RIF-induced changes in gene expression in the WT versus ΔsigB mutant are shown using genes with a q value of <0.1. (c) Complemented strains were created by integrating MSMEG_2539, MSMEG_2252, MSMEG_2254, and MSMEG_2174 (genes highly upregulated in the presence of RIF) and MSMEG_4708 and MSMEG_6241(two SigB-dependent genes identified by RNA-seq) at the Bxb1 attB site of mc2155 ΔsigB. Tenfold serial dilutions of M. smegmatis mc2155, the MsΔsigB mutant, and the complemented strains were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 ADC plates containing the indicated concentration of RIF. Overexpression of the genes listed above did not restore the RIF-sensitive phenotype of mc2155 ΔsigB. (d) Wild-type M. smegmatis and the MsΔsigB strain were grown to an A600 of 0.7 and exposed to 4 μg/ml RIF for 30 min, and the amounts of the rbpA and carD transcripts were determined by qPCR and plotted as the fold induction over the level of expression for an unexposed control. Data represent the mean ± SD (n = 3). sigA was used as an endogenous control. (e) A strain complemented with rbpA was created by integrating MSMEG_3858 at the Bxb1 attB site of mc2155 ΔsigB. Tenfold serial dilutions of M. smegmatis mc2155, the MsΔsigB mutant, and the rbpA complemented strain were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 ADC plates containing the indicated concentration of RIF.