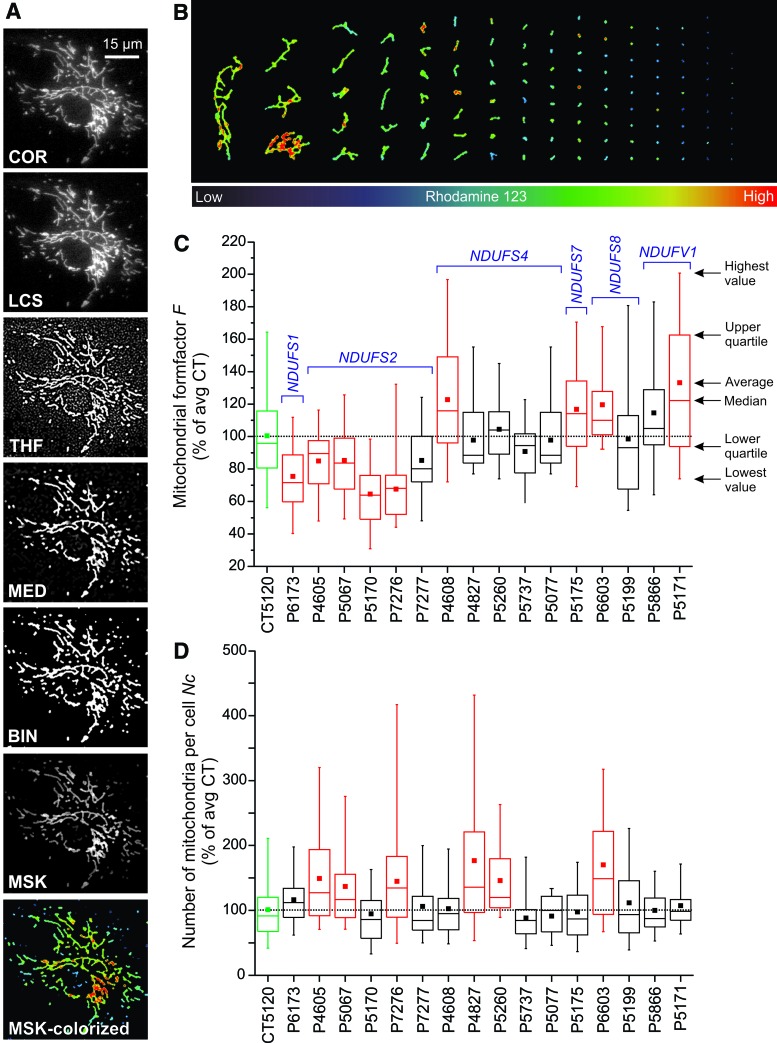
FIG. 13.
Analysis of mitochondrial morphology in fibroblasts with isolated CI deficiency by microscopy image processing and quantification. PHSFs were stained with the mitochondria-accumulating cation rhodamine 123 and visualized by using video-rate confocal microscopy (1 s image acquisition time; 30 images/s were averaged). (A) Illustration of the image processing strategy using control CT5120 cells. The acquired images were subsequently background corrected (COR), subjected to an LCS operation, processed by a THF, processed by a median (MED) filter, and thresholded to obtain a binarized (BIN) black-and-white image. The COR image was masked by the BIN image, yielding a masked (MSK) image that was colorized (MSK-colorized; blue = low and red = high fluorescence intensity). This processing strategy was described in detail elsewhere (186, 192, 194). (B) Image depicting the mitochondrial objects in the MSK-colorized image sorted according to their size (area) from left to right. This “mitogram” reveals considerable heterogeneity in rhodamine 123 fluorescence signal between and within individual mitochondrial objects. (C) Application of the strategy in (B) on primary skin fibroblasts of control subjects (CT5120; green) and patients (P) with isolated CI deficiency. The latter carried mutations in various CI subunit-encoding nDNA genes (NDUFS1, NDUFS2, NDUFS4, NDUFS7, NDUFS8, and NDUFV1). The boxplot reveals that the mitochondrial formfactor (F; a combined measure of mitochondrial length and degree of branching) is significantly altered (colorized red) or similar (black) to CT5120 cells (green). (D) Similar to (C), but now depicting the number of mitochondria per cell (Nc). The interpretation and implications of this data is presented elsewhere (187, 193, 195). LCS, linear contrast stretch; THF, top-hat filter. Color images are available online.