FIG. 14.
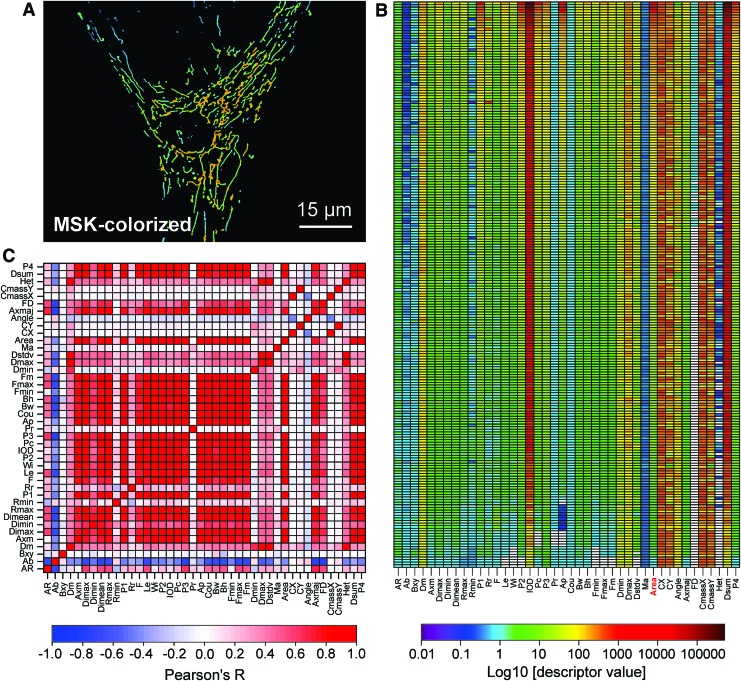
High-content analysis (“cellomics”) of mitochondrial morphofunction. PHSFs (CT5120) were stained with the mitochondria-accumulating cation TMRM and visualized by using fluorescence microscopy. The acquired images were processed as described in Figure 13A. (A) Typical TMRM fluorescence image used to illustrate the high-content analysis. (B) Heat map depicting the numerical value of 42 morphofunctional descriptors (horizontal axis; Table 2) for each of the 217 mitochondrial objects in (A) (vertical axis). These data were sorted from large (top) to small (bottom) values of mitochondrial size (descriptor “Area”; marked in red) and Log10 scaled. Individual horizontal rows represent the morphofunctional “fingerprint” for each mitochondrial object (30), which can be averaged at the level of individual cells and cell populations to obtain information at different complexity levels. (C) Degree of linear correlation (calculated by using Pearson's R) between the individual descriptors in (B). Negative and positive correlations are depicted in blue and red, respectively. This correlation plot also can be considered a cellular fingerprint of mitochondrial morphofunction in the analyzed cell. The data presented in this figure can be explored and classified by using various computational techniques (147, 411), allowing time analysis, comparison of different cells and cell types, culture conditions, and modulation strategies. Color images are available online.