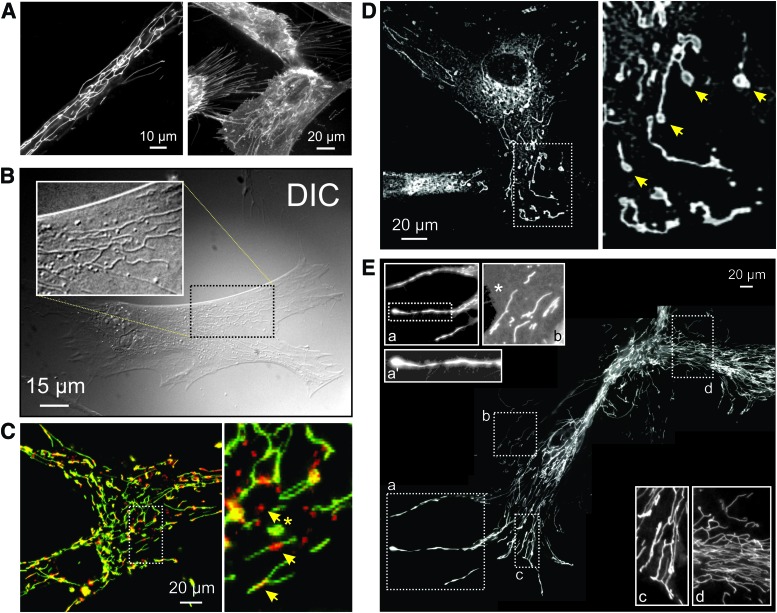
FIG. 5.
Mitochondrial morphology visualized by using live-cell fluorescence microscopy. (A) Left panel: visualization of mitochondrial structure in a primary fibroblast of a patient carrying a compound heterozygous mutation (R59X/T423M) in the gene encoding the NDUFV1 subunit of CI (left panel). Cells were stained with the mitochondria-accumulating cation rhodamine 123. Right panel: PHSFs (control CT5120) co-stained with rhodamine 123 and the membrane-staining reporter molecule FM1-43. (B) Visualization of mitochondrial structure in a PHSF (control CT5120) using DIC imaging (also known as Nomarski microscopy). Cells were cultured in the presence of the CI inhibitor rotenone (100 nM, 72 h). The white box shows a zoom-in of the indicated region. (C) Confocal image of a control PHSF stained with JC-1, which accumulates in the mitochondrial matrix according to the Δψ across the MIM. The right part of the image shows a zoom-in of the indicated region. Arrows mark regions of JC-1 aggregation (top arrow; red), suggesting a more negative Δψ, and inhomogeneous JC-1 signals across mitochondrial filaments (middle and lower arrow). (D) Microscopy image of a PHSF (control CT5120) stained with MitoTracker Green FM. This image was obtained by using video-rate microscopy and deconvolved off-line. Arrows mark “donut-shaped” mitochondria. (E) Collage of microscopy images visualizing a large mitochondrial network in rhodamine 123-stained PHSFs of a patient carrying a mutation (R228Q) in the gene encoding the NDUFS2 subunit of CI. These cells were imaged following immortalization and chromosome transfer. Magnifications (a, a', b, c, d) show mitochondrial morphologies throughout the cell. The magnified images were contrast-optimized for visualization purposes (an asterisk shows the cytosol). DIC, differential interference contrast; PHSFs, primary human skin fibroblasts. Color images are available online.