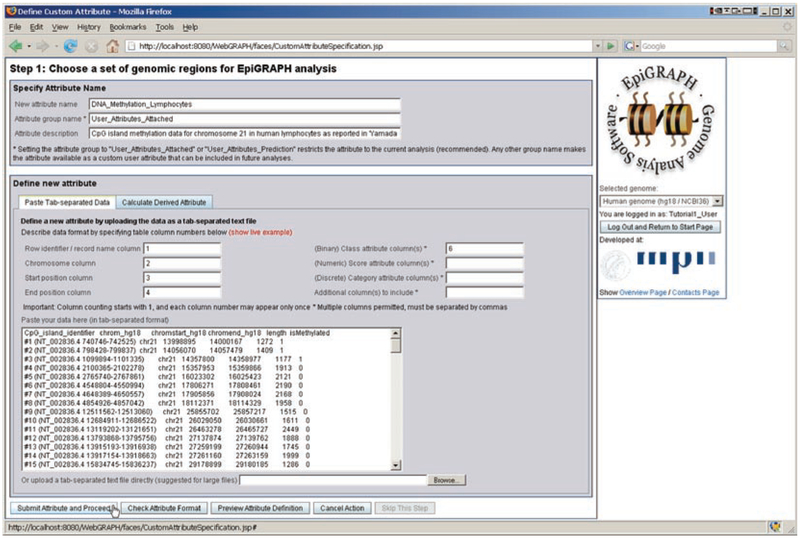
Fig. 2.
Submitting a custom dataset for analysis with EpiGRAPH. This screenshot displays EpiGRAPH’s attribute submission page, consisting of a brief attribute documentation (top), a set of text fields in which the column semantics are specified (e.g., which column contains the chromosome name and the start and end position for each genomic region) and a large text area into which a tab-separated table of genomic regions can be pasted. Due to different column widths, the columns of the table are not properly aligned, which is often the case and will not cause any problems. Importantly, each row in the table must correspond to exactly one genomic region, and its location in terms of chromosome name, start position and end position must be specified relative to the genome assembly selected in the choice box below the EpiGRAPH logo on the right of the screen (“hg18” in this case)