FIGURE 3.
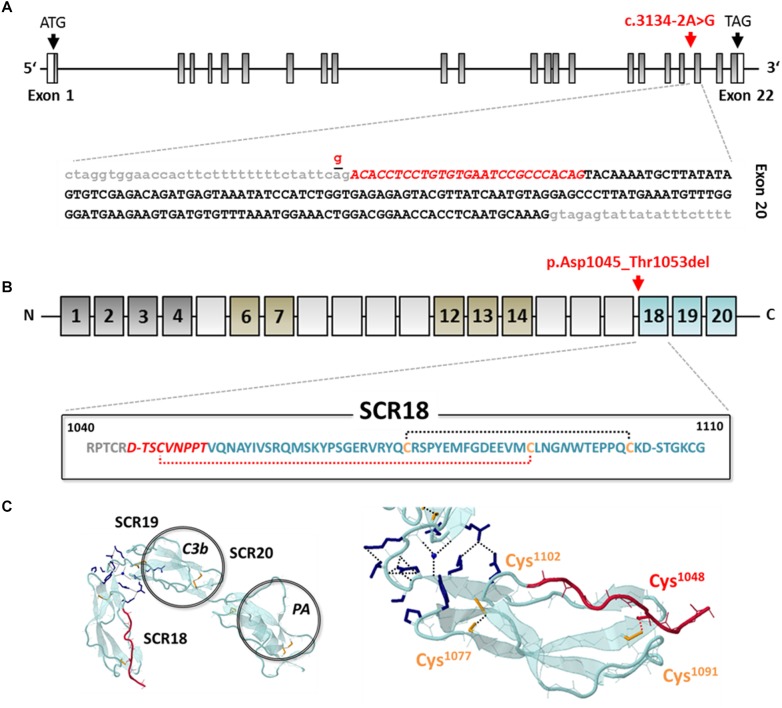
Genetic analysis and in silico evaluation of CFH c.3134-2A>G; p.Asp1045_Thr1053del. (A) Exon structure of the CFH gene indicating the position of the identified variant (red arrow and letters) at the splice acceptor site of the intron (gray letters) 19/exon (black letters) 20 boundary. The variant activates an exonic splice acceptor and leads to a partial loss of exon 20 from the RNA transcript (red italic letters). (B) Domain structure of CFH (dark gray: regulatory domain; brown: proteoglycan binding; blue: surface binding domain); the variant results in an in-frame deletion of 9 amino acids of SCR17 and SCR18 including Cys1048 (red italic letters) leading to the loss of a structure-determining disulfide bond (red-dotted line). (C) Model of the C-terminal domains SCR18-20 of CFH (PDB: 3SW0) modified from Morgan et al. (2012), indicating the deleted sequence part (red color) (Morgan et al., 2012). SCR, short consensus repeat; C3b, C3 binding site; PA, polyanion binding site.