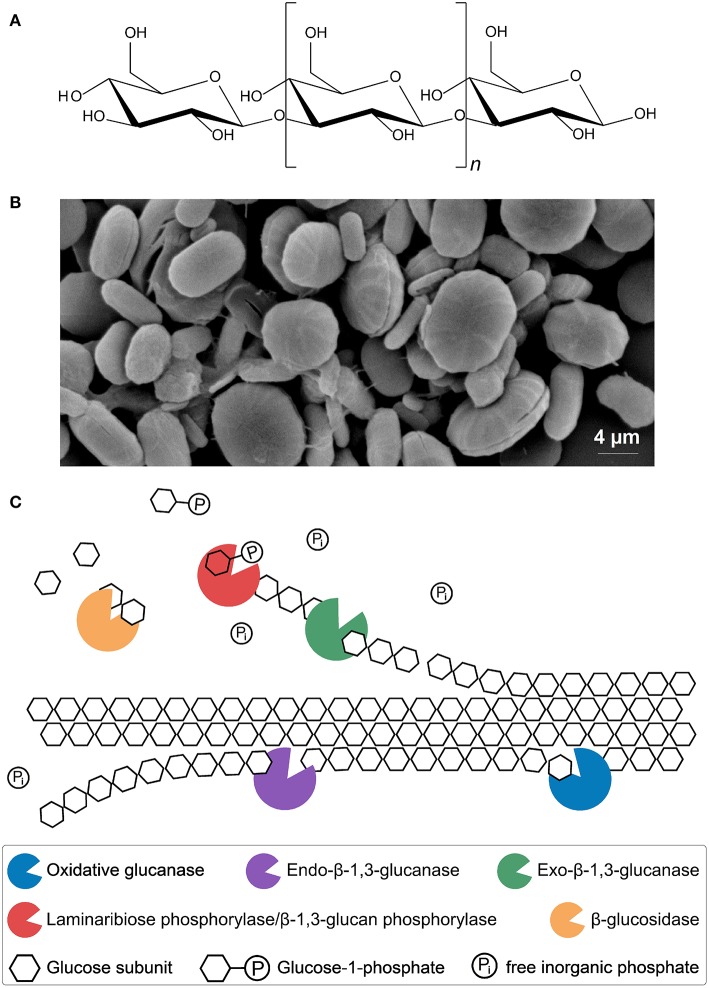
Figure 3.
Molecular structure of paramylon (A). β-1,3-glucan chain (~700 ≤ n ≥ 3,000) (Miyatake and Kitaoka, 1983; Koizumi et al., 1993; Barsanti et al., 2011). Microscopic image of paramylon granules (B). Shown are ellipses and rods without biomembrane (Bäumer et al., 2001; Monfils et al., 2011). Putative enzymatic mechanism for synergistic paramylon degradation in Euglena gracilis (C). Enzymes, substrates and products are shown. Oxidative glucanases: cleavage of crystalline paramylon to make it accessible for other enzymes, possibly similar to oxidative cleavage of cellulose (Johansen, 2016). Hydrolytic endo-β-1,3-glucanases: random cleavage of the polysaccharide chain (Takeda et al., 2015). Hydrolytic exo-β-1,3-glucanases and β-glucosidases: oligoglucans and single glucose units are cleaved off, respectively, at the ends of accessible and freed polysaccharide chains, comparable to the cellulolytic enzymes from Trichoderma reesei (Barras and Stone, 1969; Jeng et al., 2011; Keshavarz and Khalesi, 2016). Laminaribiose phosphorylase/β-1,3-glucan phosphorylase: cleaving of laminaribiose/laminarioligosaccharides into glucose/laminarioliosaccharides and glucose-1-phosphate, which requires free inorganic phosphate (Marechal, 1967; Kuhaudomlarp et al., 2018).