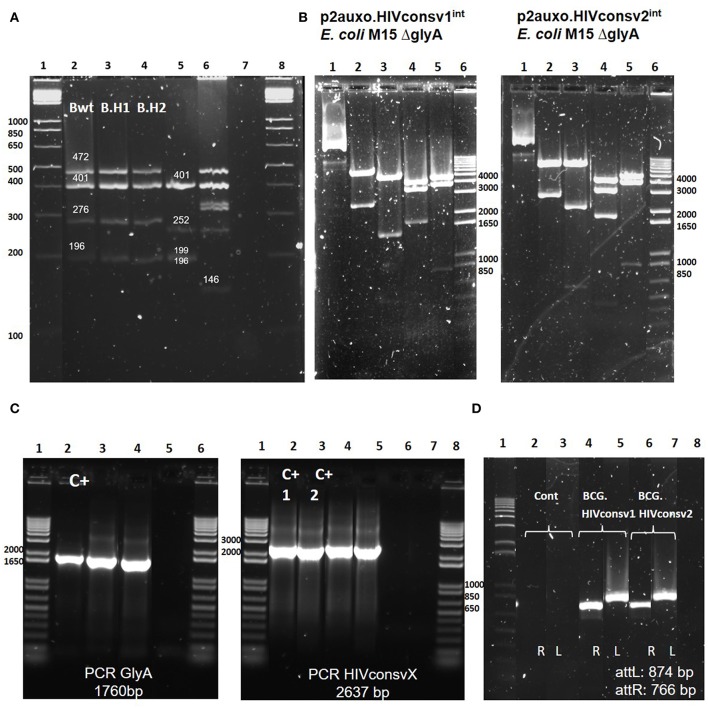
Figure 2.
Genetic characterization of the BCG.HIVconsv1&22auxo.intstrains. (A) Pasteur substrain identification of BCG.HIVconsv1&22auxo.intstrains by multiplex PCR assay. Lane 1 and 8: molecular weight marker; lane 2: BCG wild type Pasteur giving the bands of 472, 401, 276, and 196 bp; lane 3 and 4: BCG.HIVconsv1&22auxo.int Working Vaccine Stocks (WVS), respectively, giving the bands of 472, 401, 276, and 196 bp; lane 5: BCG Connaught giving the bands of 401, 252,199, and 196 bp; lane 6: MTBVAC.HIVA2auxo.int (M.tuberculosis strain) WVS giving a distinctive band of 146 bp, and lane 7: negative control, distilled water. (B) Enzymatic restriction analysis of p2auxo.HIVconsv1int (left) and p2auxo.HIVconsv2int (right) plasmids DNA purified from E. coli M15ΔglyA cultures (pre-BCG transformation). Lane 1: Undigested plasmids; Lane 2: HindIII digestion; Lane 3: PstI digestion; Lane 4: StuI digestion; Lane 5: XbaI digestion; Lane 6: Molecular weight marker. (C) PCR analysis of E. coli glyA DNA and HIVconsv1 or HIVconsv2 DNA coding sequences. Left panel: glyA PCR. Lanes 1 and 6: Molecular weight marker; Lane 2: p2auxo.Ag85B, positive control; Lane 3: BCG.HIVconsv1int (WVS); Lane 4: BCG.HIVconsv2int (WVS); Lane 5: distilled water, negative control. Right panel: HIVconsv1 and HIVconsv2 PCR. Lanes 1 and 8 corresponds to the molecular weight marker; Lanes 2 and 3: p2auxo.HIVconsv1 and p2auxo.HIVconsv2 (episomal), positive controls; Lane 4: BCG.HIVconsv1int (WVS); Lane 5: BCG.HIVconsv2int (WVS); Lanes 6 and 7: distilled water, negative controls; (D) PCR analysis of attR and attL DNA region using as template the cultures of BCG wild type, negative control (lane2 and 3), BCG.HIVconsv12auxo.int WVS (lane 4 and 5), BCG.HIVconsv22auxo.int WVS (lanes 6, and 7) and distilled water as negative control (lane 8). Lane 1 corresponds to molecular weight marker.