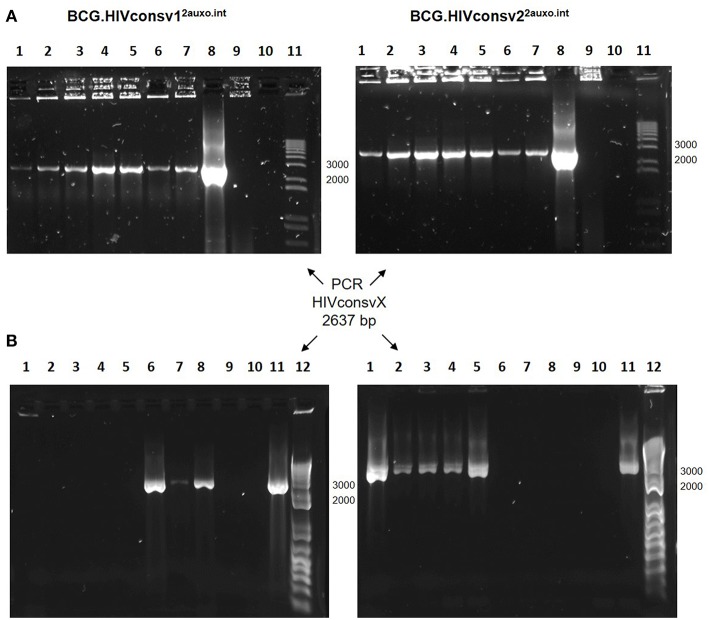
Figure 4.
Genetic stability of the BCG.HIVconsv12auxo.int and BCG.HIVconsv22auxo.int strains. (A) In vitro stability. Working vaccine stocks were subcultured in lysine-free medium for 5 weeks and PCR analysis was to confirm presence of the HIVconsv1&2 genes; BCG.HIVconsv12auxo.int (left panel) and BCG.HIVconsv22auxo.int (right panel). Lanes 1–5 correspond to weekly subcultures from week 1–5; Lanes 6&7: 1 and 2 weekly subcultures in lysine supplemented media; Lanes 8: p2auxo.HIVconsvXint plasmid (positive control); Lanes 9: distilled water (negative control); Lanes 10: BCG wt (negative control); Lanes 11: molecular weight marker. The HIVconsv1&2 genes were detected in both BCG.HIVconsv12auxo.int and BCG.HIVconsv22auxo.int strains after 5 weekly subcultures. (B) In vivo stability. Mice were injected i.d. with 106 cfu of BCG.HIVconsv1&22auxo.int and boosted i.m. with 108 IU of ChAdOx1.HIVconsv5&6, spleens were homogenized 8 weeks after BCG inoculation and the recovered rBCG colonies were tested by PCR analysis using primers specific for HIVconsv1 (left panel) or HIVconsv2 (right panel) DNA coding sequences. Representative assessment of positive colonies. Lanes 1 to 8: rBCG.HIVconsvX2auxo.int colonies recovered from the non-lysine supplemented plate; Lanes 9: BCG wt (negative control); Lanes 10: Distilled water (negative control); Lanes 11: p2auxo.HIVconsv1int or p2auxo.HIVconsv2int, respectively (Plasmid DNA positive control); Lanes 12: Molecular weight marker.