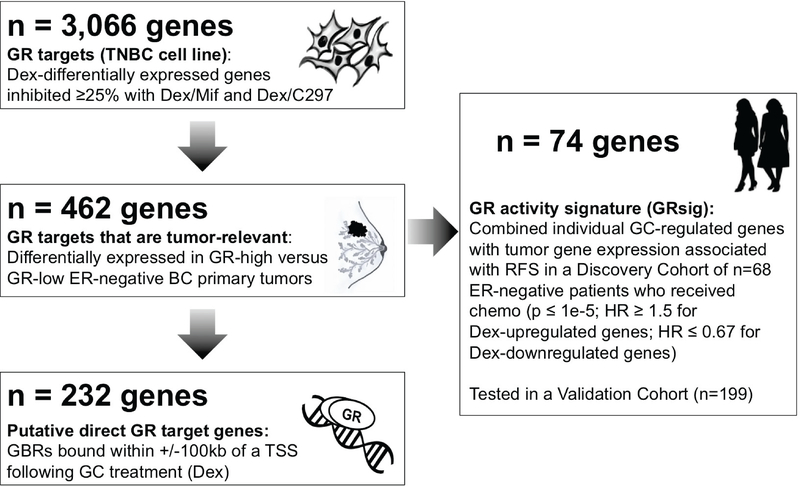
Figure 5. Identification schema for the GR activity signature (GRsig).
Genes that were Dex-regulated and inhibited at least 25% by Mif and C297 were identified in MDA-MB-231 cells (n=3,066). Next, the subset of genes also differentially expressed in the same direction in GR-high versus GR-low ER- negative BCs was identified (n=462). GR ChIP-seq determined putative GR direct target genes as having GR associated within 100kb of the gene TSS (n=232). A GR “activity signature” (GRsig) was identified based on their univariate association with RFS (HR ≥1.5 or HR ≤ 0.67; and p ≤ 1e-5) in the Discovery cohort of early-stage ER-negative BC patients with adjuvant chemotherapy. The GRsig comprised of n=74 genes which 1) included genes that were associated with poor RFS (HR ≥ 1.5) and were Dex- upregulated and 2) genes that were associated with improved RFS (HR ≤ 0.67) and were Dex- downregulated. This GRsig was applied to the Discovery and an independent Validation cohort of early- stage patients treated with adjuvant chemotherapy.