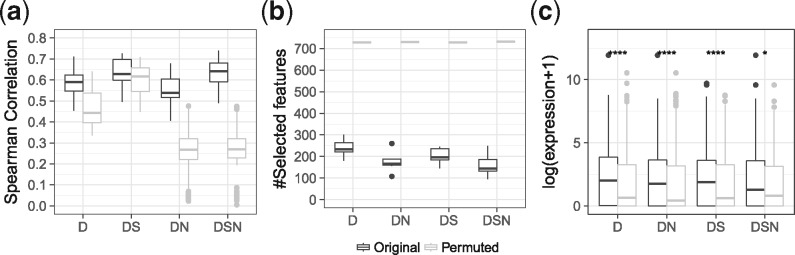
Fig. 5.
(a) This illustrates the performance of gene expression models based on four different annotation setups (D, DS, DN, DSN) for original and permuted data. Part (b) shows the number of selected features for all annotation variants in original and permuted data using elastic net regularization. Part (c) compares the expression of TFs selected by the individual models per setup against the expression of TFs selected on permuted data. According to a Wilcoxon test, the expression differences are significant in all cases with P < 0.05 denoted by *