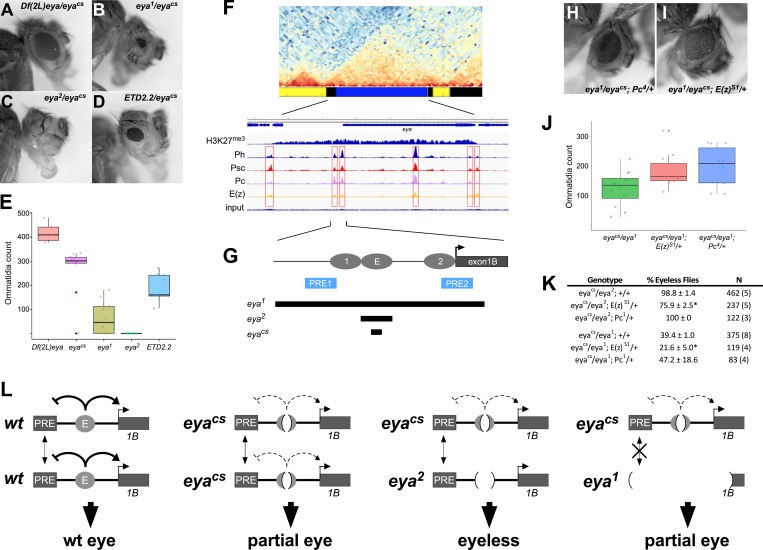
Fig 8. Pairing sensitive silencing between Class A alleles reflects a direct role for PcG proteins in regulating eya.
A-D, eyes of representative flies carrying the indicated genotypes. ETD2.2 is a transvection-disrupting rearrangement of a second chromosome carrying eya2. E, quantification of eye development for flies carrying the indicated alleles in trans to eyacs. Consistent with pairing-sensitive silencing, decreased eye development is observed when eyacs is placed in trans to other Class A alleles relative to eyacs homozygotes, and the decrease is disrupted by the eya2 rearrangement ETD2.2 (compare column 4 to column 5). Furthermore, 98.8% of eyacs/eya2 flies are completely eyeless (n = 462; see Panel K), but all (n = 120) eyacs/ETD2.2 flies develop scorable eye tissue. F, genomic features of the eya locus (note that genomic maps are reversed relative to the reference sequence.) Top, Hi-C contact map showing TAD structure, with chromatin color map below. The eya locus occupies a TAD that is primarily blue chromatin, indicative of Polycomb silencing. Below, ChIP-seq peaks for K27-trimethylated histone H3 and individual PcG proteins as assayed from larval disc tissues. Six putative PREs (red boxes) are indicated by the pattern of peaks. G, schematic showing positions of eye-specific enhancers of putative PREs upstream of the eya B promoter. Bars below the schematic indicate the deletions carried by Class A alleles. H-I, eyes of representative flies carrying the indicated genotypes. J, quantification of eye development for flies carrying the indicated genotypes. Completely eyeless flies were not scored in this analysis. Significant increases in the ommatidia count are observed for eyacs/eya1; E(z)S1/+ (p = 0.03, Mann-Whitney test) and eyacs/eya1; Pc4/+ (p = 0.04) relative to eyacs/eya1; +/+. K, scoring of flies that have at least one eye of any size in the indicated genotypes. Asterisks indicates significant difference relative to eyacs/eya2; +/+ (p = 0.008, Mann-Whitney U test) or eyacs/eya1; +/+ (p = 0.03). “N” indicates the number of flies scored, with the number of separate vials in parentheses. L, Models for interactions between eye-specific enhancers and putative PREs. For simplicity, only the E enhancer (required for initiating eye development) and its neighboring PRE are shown; we expect that other local PREs behave similarly. In wild type, we propose that the enhancer has dual roles to block silencing by PREs through an unknown mechanism and to activate transcription primarily from the eya-B promoter. The eyacs allele retains partial enhancer activity, whereas the eya2 allele has no detectable enhancer activity; in eyacs/eya2 flies, pairing between homologous PREs (dual arrows) increases their capacity to silence, resulting in an eyeless phenotype in the paired state, but a partial eye when unpaired by chromosomal rearrangements. Since eya1 deletes some of the putative PREs, pairing-dependent silencing is reduced, and the eyacs/eya1 phenotype is less severe than that of eyacs/eya2.