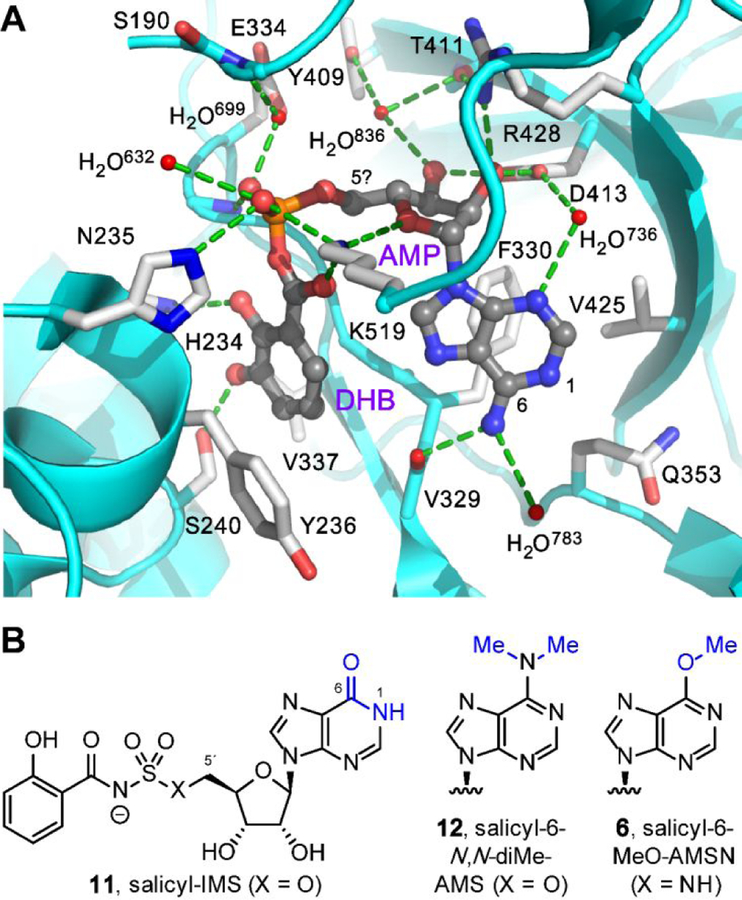
Figure 5.
Design rationales for new salicyl-AMS analogues. (A) Cocrystal structure of 2,3-dihydroxybenzoyl-AMP (DHB-AMP, gray) bound in the active site of the DHBA adenylation enzyme Bacillus subtilis DhbE (cyan and white, PDB: 1MDB),64 illustrating pharmacophoric cisoid binding conformation.65 Salicyl-AMSNMe (4b) is designed to promote this cisoid binding conformation. (B) Structures of previously reported inosine analogue salicyl-IMS (11) and dimethylated analogue salicyl-6-N,N-diMe-AMS (12), both of which are weak MbtAtb inhibitors,23 presumably due to loss of hydrogen-bonding interaction with the main-chain carbonyl oxygen of MbtAtb V352 (DhbE V329). Salicyl-6-MeO-AMS (6) is designed to remove this hydrogen-bonding interaction without introducing a proton at N1 via tautomerization (cf. 11) and without introducing a potential steric clash with V352 (cf. 12).