Figure 2.
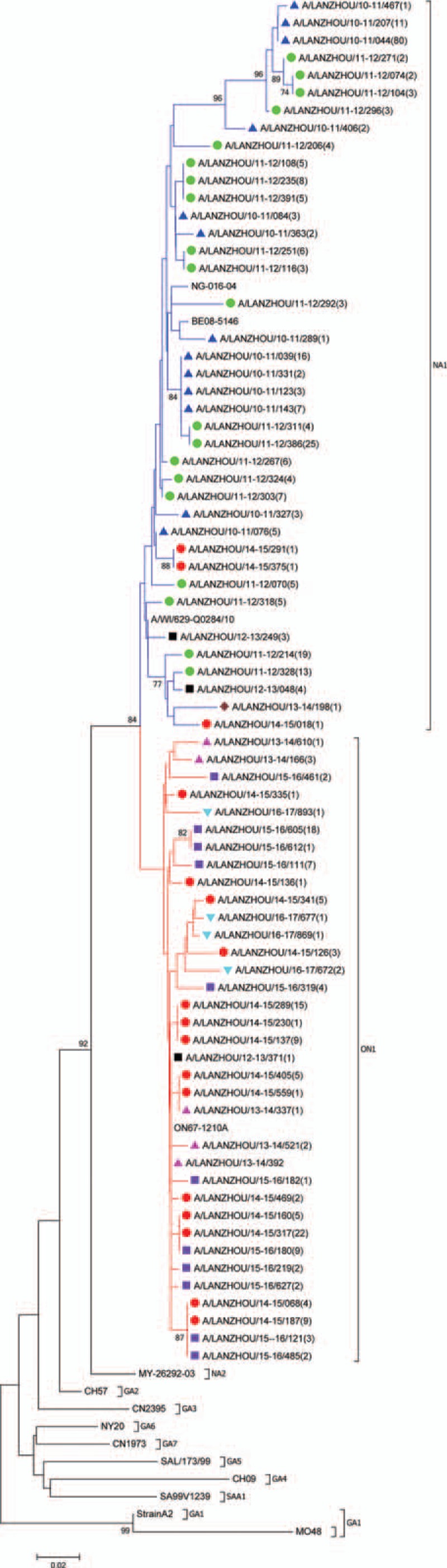
Phylogenetic tree based on the second hypervariable region of the G protein gene, Lanzhou December 2010 to June 20137 (n = 75 unique Lanzhou sequences). Multiple sequences alignment and phylogenetic trees were constructed using Clustal W and neighbor-joining algorithm running within MEGA 6 software. The scale bar shows the proportions of nucleotide substitutions per site. Numbers at nodes are bootstrap values for 1000 iterations; only bootstrap values of >70% are shown. Numbers in round brackets indicate the total number of strains with an identical sequence. Reference strains representing known genotypes are indicated in bold. Branches are color-coded according to the deduced amino acid sequence, identifying subtrees and genotypes—red: sequences with insertion clustering with the novel ON1 genotype; blue: sequences of NA1 genotype. Symbol color indicates epidemic season—blue: 2010/2011; green: 2011/2012; black: 2012/2013;brown: 2013/2014; red: 2014/2015; purple 2015/2016; turquoise blue: 2016/2017.
