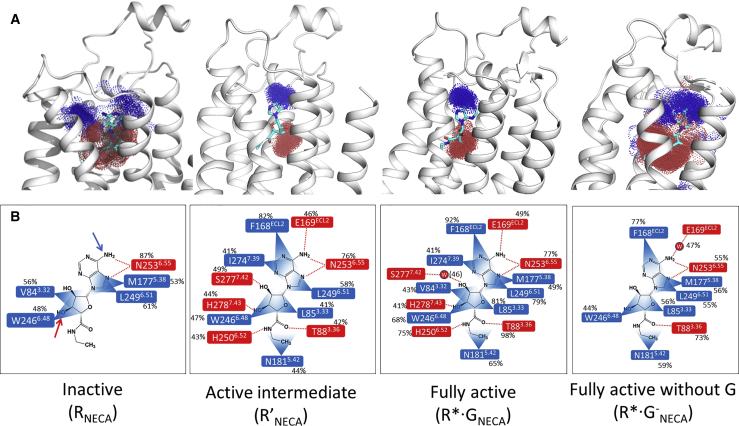
Figure 3.
Mobility and Binding of NECA in Different Conformational States
(A) Spatial distribution function of the agonist NECA calculated centering on the nitrogen atom from the primary amine group and the oxygen atom of the hydroxyl group in the sugar ring of NECA, blue and red arrows in (B) (see Figure S3 for data on adenosine).
(B) The protein-ligand contacts for NECA binding in RNECA, R′NECA, R∗·GNECA, and R∗·G−NECA states of A2AR. The protein-ligand contacts that are polar are marked in red and hydrophobic residue contacts are shown in blue. The percentage of snapshots within the MD simulations for each of these protein-ligand contacts is given (aggregated trajectory of 1 μs, 50,000 snapshots per calculation). N253 makes hydrogen bonds with two different N atoms on the adenine ring and the percentage shown is the sum of both.