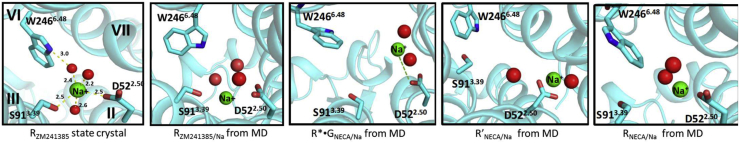
Figure 6.
Effect of Na+ Ions in Diverse Conformational States
Effect of Na+ ion in the MD simulations of the inverse agonist ZM241385-bound inactive state R (RZMA241385/Na), agonist NECA-bound inactive state R (RNECA/Na), active-intermediate state R′ (R′NECA/Na), and G protein-bound fully active state R∗·G (R∗·GNECA/Na). The Na+ ion retains the hydrogen bonds with residues D522.50 and S913.39 and a water-mediated hydrogen bond with W2466.48 during the MD simulations of ZM241385-bound R state and as seen in the crystal structure of ZM241385-bound inactive state of A2AR (PDB: 4EIY). Agonist NECA binding in the R′ and R∗·G states disrupts the interaction of the Na+ with S913.39 but retains the interaction with D522.50. The waters present in the ZM241385-bound R state rearrange in the agonist-bound simulations.