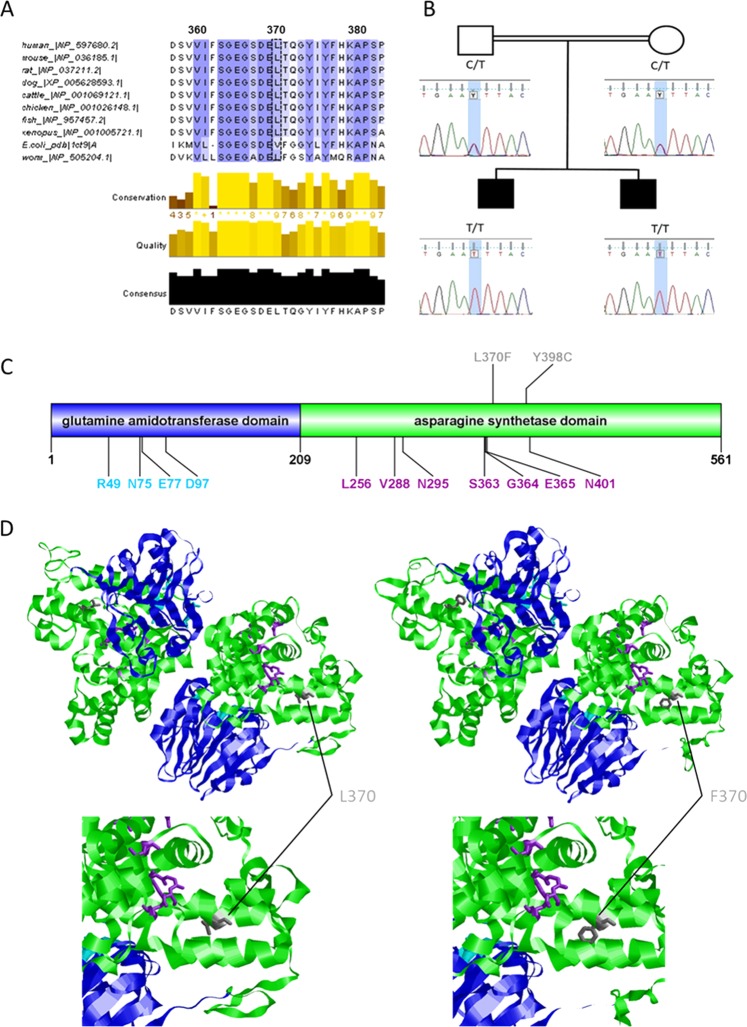
Fig. 2. Conservation, cosegregation, and consequences of the mutated residue p.Leu370Phe.
a Multiple alignment of human ASNS (NP_597680.2) with selected orthologues of mouse (NP_036185.1), rat (NP_037211.2), dog (XP_005628593.1), cattle (NP_001069121.1), chicken (NP_001026148.1), fish (NP_957457.2), Xenopus (NP_001005721.1), Escherichia coli (PDB 1CT9.1. A), and worm (NP_505204.1) shows that a highly conserved amino acid region is affected. Amino acid color labels were selected for the block substitution matrix 62. b Pedigree and chromatograms of the DNA sequence changes observed in the ASNS gene. Sanger sequencing revealed that both parents harbor a heterozygous variant at position c.1108C>T in ASNS (NM_133436.3). Both children are homozygous for this variant. c Linear model of the structure of the human ASNS protein. Blue: N-terminal domain containing the binding pocket for glutamine. Green: C-terminal domain containing the ATP-binding site. Cyan: Residues forming the glutamine-binding pocket. Purple: Residues for ATP binding through hydrogen bonds2. Gray: Localization of amino acid changes in reported patients who received oral asparagine supplementation4,6. d 3D structure model of ASNS, including wild type (UniProt ID P08243) and mutant p.Leu370Phe, by SWISS-MODEL using the crystal structure of asparagine synthetase B from Escherichia coli (PDB 1CT9.1.A)25. Gray: The affected amino acid p. Leu370 and the mutated amino acid p.Phe370 are located in the asparagine synthetase domain. Cyan: Residues p.Arg49, p.Asn75, p.Glu77, and p.Asp97 are important for glutamine binding. Purple: Amino acids p.Leu256, p.Val288, p.Asn295, p.Ser363, p.Gly364, and p.Glu365 are proposed as representing a binding site for ATP through hydrogen bonds2.