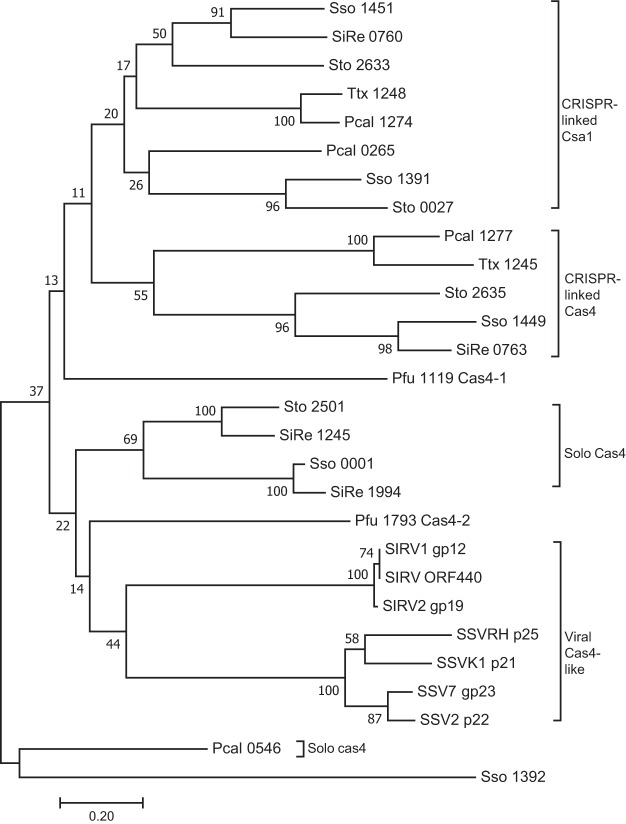
FIG 1.
Unrooted bootstrapped phylogenetic tree showing a subset of archaea and their virus-encoded Cas4 family proteins. Each protein is represented by a species code followed by the gene number (Sso, S. solfataricus; SiRe, S. islandicus REY15A; Pfu, P. furiosus; Sto, Sulfolobus tokodaii; Ttx, Trichomonas tenax; Pcal, Pyrobaculum calidifontis; SIRV, Sulfolobus islandicus rod-shaped virus; SSV, Sulfolobus spindle-shaped virus). This neighbor-joining tree was generated from a T-Coffee alignment of the proteins using Mega7, with pairwise distances between sequences uncorrected. The bootstrap values shown at each node represent the percentage of all trees (10,000 total) agreeing with this topology. The subgroups of CRISPR-associated Csa1, CRISPR-associated Cas4, solo Cas4, and viral Cas4 like proteins are indicated.