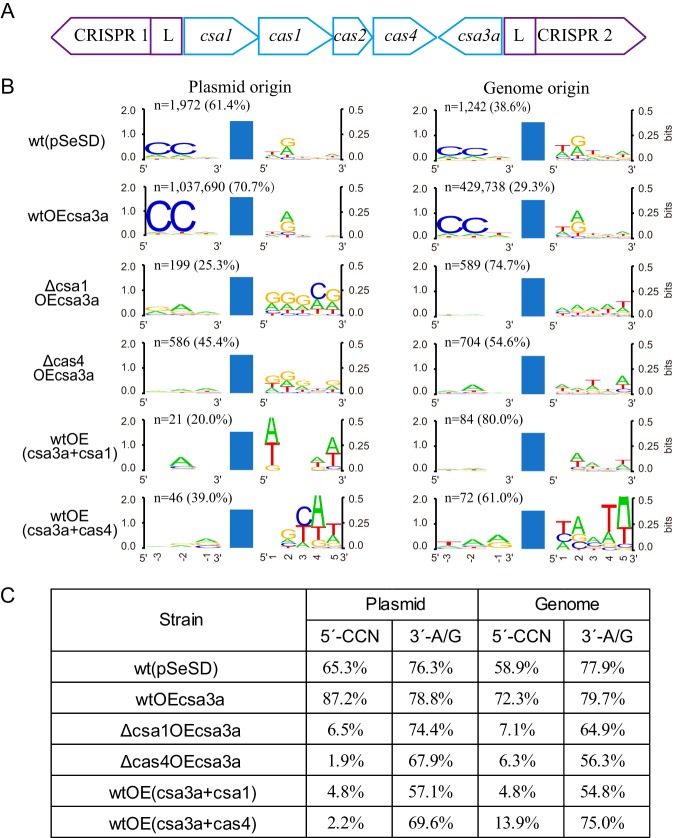
FIG 2.
Effects of Cas4 proteins on the spacer origin, PAM, and 3′ conserved motif. (A) Organization of the CRISPR-Cas adaptation module of S. islandicus REY15A with two CRISPR arrays. (B) WebLogo analysis of the conserved motifs upstream and downstream of the protospacers detected in different Sulfolobus transformants by high-throughput sequencing data. Reads of the new spacers (n) and the percentage of plasmid- or genome-derived spacers are indicated. wt(pSeSD), wild-type cells carrying empty vector; wtOEcsa3a, wt cells overexpressing the csa3a activator gene; Δcsa1OEcsa3a, csa1 deletion strain overexpressing csa3a; Δcas4OEcsa3a, cas4 deletion strain overexpressing csa3a; wtOE(csa3a+csa1), wt cells overexpressing both csa3a and csa1; wtOE(csa3a+cas4), wt cells overexpressing both csa3a and cas4. Blue bars, protospacers. (C) Percentage of the new spacers of plasmid or genome origin carrying CCN or A/G motifs from the different genetically modified strains.