Pseudomonas aeruginosa is often found with bacteria and fungi that produce fermentation products, including ethanol. At concentrations similar to those produced by environmental microbes, we found that ethanol stimulated expression of trehalose-biosynthetic genes and cellular levels of trehalose, a disaccharide that protects against environmental stresses. The induction of trehalose by ethanol required the alternative sigma factor AlgU through DksA- and SpoT-dependent (p)ppGpp. Trehalose accumulation also required AHL quorum sensing and occurred only in post-exponential-phase cultures. This work highlights how cells integrate cell density and growth cues in their responses to products made by other microbes and reveals a new role for (p)ppGpp in the regulation of AlgU activity.
KEYWORDS: (p)ppGpp, AlgU, DksA, Pseudomonas aeruginosa, SpoT, ethanol, microbe-microbe interaction, quorum sensing, trehalose
ABSTRACT
Pseudomonas aeruginosa frequently resides among ethanol-producing microbes, making its response to the microbially produced concentrations of ethanol relevant to understanding its biology. Our transcriptome analysis found that genes involved in trehalose metabolism were induced by low concentrations of ethanol, and biochemical assays showed that levels of intracellular trehalose increased significantly upon growth with ethanol. The increase in trehalose was dependent on the TreYZ pathway but not other trehalose-metabolic enzymes (TreS or TreA). The sigma factor AlgU (AlgT), a homolog of RpoE in other species, was required for increased expression of the treZ gene and trehalose levels, but induction was not controlled by the well-characterized proteolysis of its anti-sigma factor, MucA. Growth with ethanol led to increased SpoT-dependent (p)ppGpp accumulation, which stimulates AlgU-dependent transcription of treZ and other AlgU-regulated genes through DksA, a (p)ppGpp and RNA polymerase binding protein. Ethanol stimulation of trehalose also required acylhomoserine lactone (AHL)-mediated quorum sensing (QS), as induction was not observed in a ΔlasR ΔrhlR strain. A network analysis using a model, eADAGE, built from publicly available P. aeruginosa transcriptome data sets (J. Tan, G. Doing, K. A. Lewis, C. E. Price, et al., Cell Syst 5:63–71, 2017, https://doi.org/10.1016/j.cels.2017.06.003) provided strong support for our model in which treZ and coregulated genes are controlled by both AlgU- and AHL-mediated QS. Consistent with (p)ppGpp- and AHL-mediated quorum-sensing regulation, ethanol, even when added at the time of culture inoculation, stimulated treZ transcript levels and trehalose production in cells from post-exponential-phase cultures but not in cells from exponential-phase cultures. These data highlight the integration of growth and cell density cues in the P. aeruginosa transcriptional response to ethanol.
IMPORTANCE Pseudomonas aeruginosa is often found with bacteria and fungi that produce fermentation products, including ethanol. At concentrations similar to those produced by environmental microbes, we found that ethanol stimulated expression of trehalose-biosynthetic genes and cellular levels of trehalose, a disaccharide that protects against environmental stresses. The induction of trehalose by ethanol required the alternative sigma factor AlgU through DksA- and SpoT-dependent (p)ppGpp. Trehalose accumulation also required AHL quorum sensing and occurred only in post-exponential-phase cultures. This work highlights how cells integrate cell density and growth cues in their responses to products made by other microbes and reveals a new role for (p)ppGpp in the regulation of AlgU activity.
INTRODUCTION
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium that can cause acute and chronic infections in a broad range of hosts. P. aeruginosa frequently causes chronic infections in individuals with the genetic disorder cystic fibrosis (CF). Diverse bacterial and fungal taxa often coinfect with P. aeruginosa in CF airways (1–5), and many of these taxa are robust fermenters capable of ethanol (EtOH) production (6, 7). Ethanol has also been identified as a volatile biomarker in exhaled breath condensates that discriminates between healthy individuals and those with CF (8).
Ethanol has a range of biological activities that vary based on its concentration. At concentrations in the 3% to 5% range and higher, ethanol can inhibit growth or kill P. aeruginosa (9–11). The effects of biologically produced concentrations of ethanol within the range experienced by organisms in polymicrobial communities (0.1 to 1.1%) (12–20) have been less well studied. Several studies have shown that 1% ethanol can alter pathogenesis and interspecies interactions (12, 14, 16, 18). In Acinetobacter baumannii, ethanol enhances virulence toward Caenorhabditis elegans (16) and Galleria mellonella (18). This may be due to enhanced production of cytotoxic phospholipase C and increased expression of nutrient uptake pathways (17). In P. aeruginosa, ethanol produced by Candida albicans influences the expression of the antifungal phenazine 5-methyl phenazine-1-carboxylic acid (5MPCA), and 1% ethanol was sufficient to modulate phenazine production and stimulate exopolysaccharide Pel- and Psl-mediated biofilm and pellicle formation (12, 15).
The P. aeruginosa alternative sigma factor AlgU, also named AlgT, has been well studied for its positive regulation of alginate, an exopolysaccharide (21, 22). AlgU is an extracytoplasmic sigma factor that is homologous to RpoE (σE or σ22) in other Gram-negative bacteria (23). P. aeruginosa mutants lacking algU have increased resistance to hydrogen peroxide compared to their alginate-overproducing mucoid counterparts due to transcriptional derepression of the catalase gene katA but are more susceptible to host antimicrobial peptides (24, 25). In other species, σE is necessary for fitness in response to high concentrations of ethanol (3% to 10%) and inhibitory concentrations of salt (NaCl) (26–29). A well-known mechanism of σE activation in response to stresses that perturb the cell envelope is by proteolytic degradation of its anti-sigma factor by specific proteases (30, 31). In P. aeruginosa, the AlgU anti-sigma factor is MucA, and mucA mutations lead to high AlgU activity. Naturally occurring mucA mutants are frequently observed in populations from chronic P. aeruginosa lung infections, and these strains overproduce alginate (32, 33).
In E. coli and Salmonella enterica, σE activity can also be modulated by the alarmone (p)ppGpp (34, 35). (p)ppGpp is an intracellular molecular signal that is synthesized by either the synthase RelA or a hybrid synthase/hydrolase, SpoT (36), in response to nutrient limitation and various environmental stressors (37). (p)ppGpp can complex with the RNA polymerase binding protein DksA to promote transcription initiation and elongation and alter the effects of RNA polymerase-associated sigma factors, including RpoE (38, 39).
In this work, we show that a subinhibitory concentration of ethanol (1%) induces the expression of genes involved in trehalose metabolism and that biochemical assays detect significant increases in intracellular trehalose, a disaccharide that serves as both a compatible solute and a carbon source. Increased trehalose in response to ethanol required the TreYZ trehalose-biosynthetic enzymes but not the TreS trehalose synthase. Ethanol induction of treZ gene expression and trehalose requires the sigma factor AlgU. AlgU was activated not by release from MucA but rather in a manner dependent on (p)ppGpp. Ethanol caused a 2.5-fold increase in (p)ppGpp levels, which was specifically dependent on the SpoT (p)ppGpp synthase. The (p)ppGpp-binding protein DksA was required for ethanol-induced stimulation of treZ gene expression and trehalose levels. Consistent with previous reports (40, 41), acylhomoserine lactone (AHL)-mediated quorum sensing (QS) was also required for transcriptional induction of the treZ gene by ethanol, as a ΔlasR ΔrhlR mutant defective in AHL-mediated quorum sensing did not show increased trehalose levels in response to ethanol. The stimulation of trehalose levels by NaCl required AlgU, and trehalose stimulation in response to NaCl was lower, but still occurred, in mutants lacking the factors necessary for the response to ethanol. Analysis of genes differentially expressed when ethanol is in the growth medium, performed using the gene expression model eADAGE (ensemble analysis using denoising autoencoders of gene expression), constructed with data from over 1,056 different samples (42), placed the treYZ genes among a cluster of coregulated genes within the AHL-controlled QS and AlgU regulons. Ethanol, even when added at the time of culture inoculation, stimulated AlgU-regulated genes and trehalose production in cells only during the post-exponential phase, which is consistent with our model, in which regulators that monitor growth and cell density cues are integrated into the P. aeruginosa response to ethanol.
RESULTS
Analysis of the transcriptome upon growth in the presence of ethanol.
To examine the transcriptional response of P. aeruginosa to 1% ethanol, RNA from P. aeruginosa grown as colony biofilms for 16 h on tryptone agar with or without 1% ethanol was analyzed using P. aeruginosa Affymetrix GeneChips, as we have previously shown that ethanol increases Pel exopolysaccharide production under similar conditions (12). Similar to published results (12), the presence of 1% ethanol in the medium did not affect the number of CFU in colony biofilms (see Fig. S1A in the supplemental material).
Levels of 54 transcripts were higher by 2-fold or more in cells grown with ethanol, with a false-discovery rate (FDR)-corrected P value of less than 0.05, and the levels of 20 genes were found to be lower by 2-fold or more in the presence of ethanol (see Table S3A and B in the supplemental material). Among the most differentially expressed genes were those involved in trehalose metabolism: treZ (3-fold), treA (2.1-fold), and treS (2.5-fold) (43). Other genes that were changed upon growth with ethanol are discussed in more detail below. To determine if ethanol also led to increased levels of trehalose, intracellular trehalose concentrations were measured in cells grown with or without ethanol in lysogeny broth (LB), a nutrient-rich medium, and M63, a defined medium. The LB was pH buffered with HEPES because we observed that ethanol led to a lower final pH in P. aeruginosa cultures grown with ethanol (final pH 8.3 in the control and pH 6.5 in ethanol from an initial pH 7.1), despite similar growth kinetics (see Fig. S1B), as has been described for Escherichia coli (44) and A. baumannii (18). Ethanol led to significant increases in trehalose in both buffered LB (>2-fold higher with ethanol) and M63 (20-fold higher with ethanol).
Increased trehalose in response to ethanol requires treYZ genes.
In pseudomonads, such as P. aeruginosa and Pseudomonas syringae, trehalose can be synthesized by the TreYZ pathway, which converts glycogen to trehalose via a maltooligosyl trehalose synthase and glycosyl hydrolase (45), and by TreS, a trehalose synthetase that uses maltose as a substrate (Fig. 1A shows a schematic) (43, 46). Trehalose is degraded by the trehalase TreA; in other species, TreS also has trehalose-catabolic activities (47, 48).
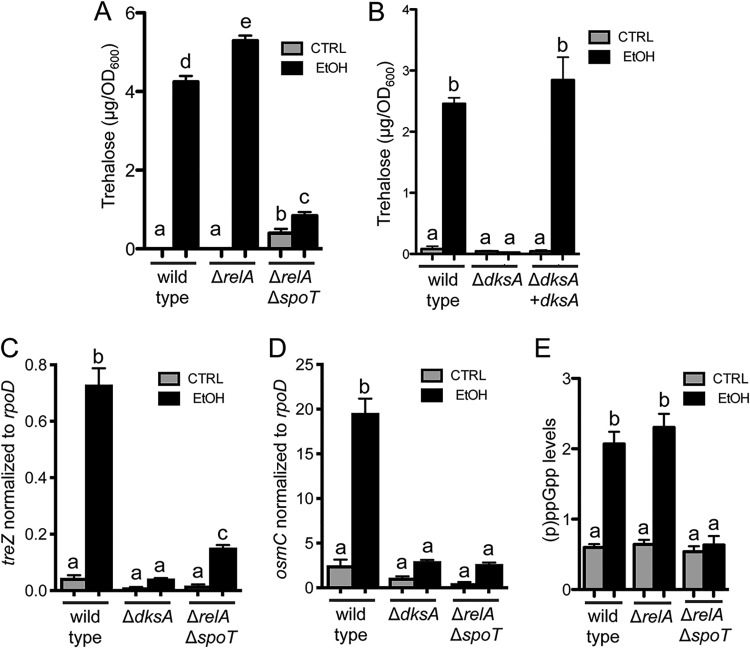
FIG 1.
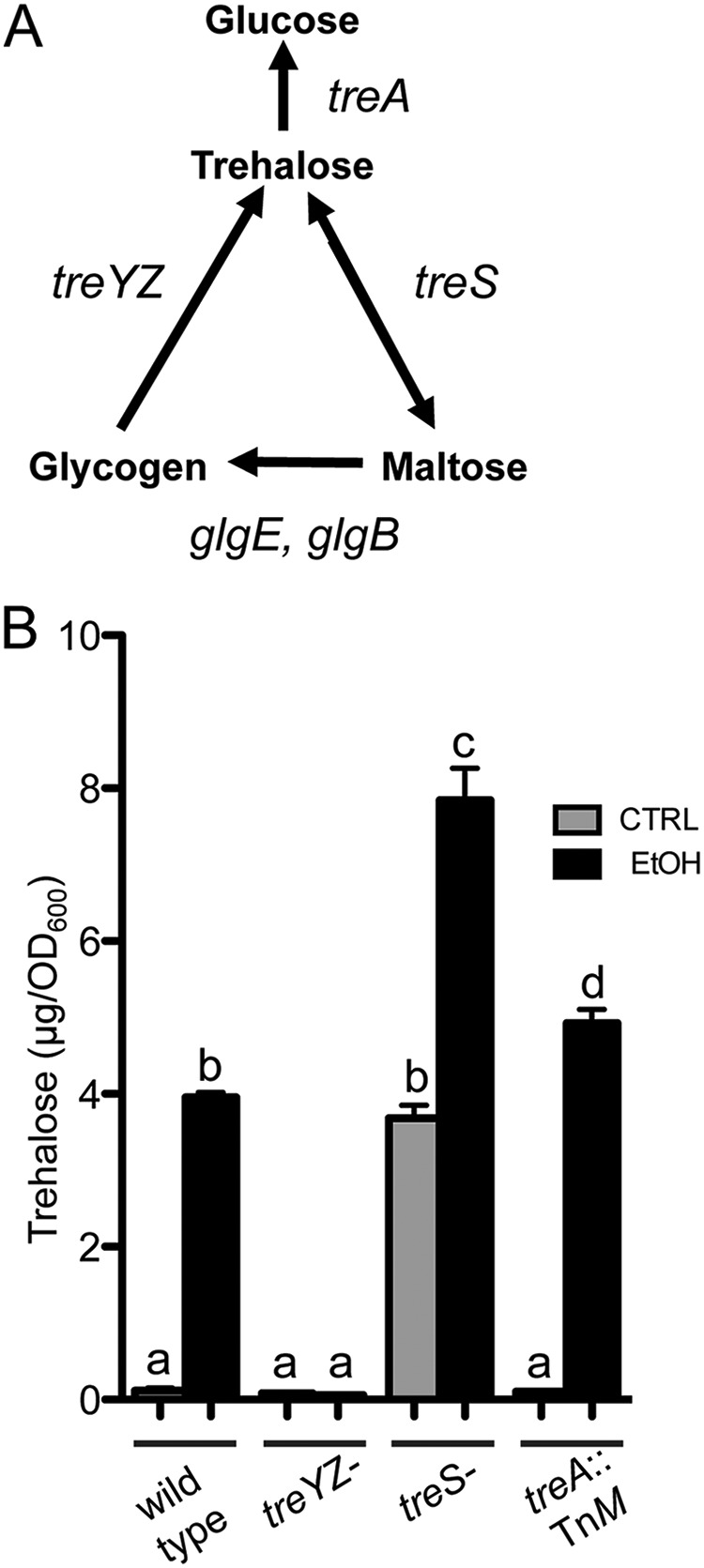
Trehalose accumulation in response to ethanol requires treYZ. (A) Schematic of trehalose-biosynthetic pathways in P. aeruginosa. (B) Trehalose levels in trehalose-metabolic mutants grown in M63 medium with and without 1% ethanol for 16 h. The data are representative of the results of at least 3 independent experiments with 3 biological replicates each. a-b, a-c, a-d, b-c, and c-d, P < 0.0001; b-d, P < 0.02 (based on two-way ANOVA and Tukey’s multiple-comparison test). The error bars indicate standard deviations.
To determine which metabolic pathway was responsible for increased trehalose in cells grown with ethanol, we used mutants lacking the 6-gene operon (PA14_36570 to PA14_36630) that contains treY and treZ genes (referred to here as a treYZ mutant [43]), the 3-gene operon that contains treS (referred to here as a treS mutant [43]), and an insertional mutant of treA (43, 49). While the treS mutant and treA::TnM strain both showed a marked increase in trehalose in the presence of ethanol in comparison to controls, the treYZ mutant strain did not, suggesting the increase in trehalose was by the TreYZ pathway (Fig. 1B). To confirm that this was due to loss of the TreYZ pathway functionality, we confirmed these results in a treZ::TnM mutant (see Fig. S2 in the supplemental material). The significantly higher levels of trehalose in the treS mutant relative to the wild type under both control and ethanol conditions suggested a role for TreS in trehalose catabolism in these cultures (50).
Ethanol catabolism occurs by the pyrroloquinoline quinone (PQQ)-dependent alcohol dehydrogenase ExaAB, and the resultant acetaldehyde is catabolized through a pathway that includes acetyl coenzyme A (acetyl-CoA) synthetase (AcsA). We have shown previously that ethanol-catabolic (exaA, pqqB, and acsA) mutants are defective in growth on ethanol as a carbon source (12). Ethanol-catabolic mutants still showed stimulation of trehalose production with ethanol in the growth medium (see Fig. S3A in the supplemental material).
Because we previously showed that ethanol (1%) led to increased production of the Pel exopolysaccharide through the diguanylate cyclase WspR (12), we determined if changes in trehalose occurred in response to changes in Pel production. We found that both the pelA and wspR mutants still had higher levels of trehalose in cells grown with ethanol (see Fig. S3B), suggesting that changes in exopolysaccharide biosynthesis did not cause the increase in trehalose.
Ethanol induction of treZ gene expression and trehalose levels is dependent on AlgU.
Several lines of evidence led us to hypothesize that the alternative sigma factor AlgU controlled the induction of trehalose-metabolic genes in response to ethanol. First, treZ, treS, and treA have been reported to be differentially expressed in algU mutant strains compared to a wild-type strain in transcriptomics studies (51, 52). Second, osmC and pfpI, two well-characterized members of the AlgU regulon (52–54), were differentially expressed in our transcriptomics analysis of cells grown with or without 1% ethanol (see Table S3A).
In wild-type cells, quantitative real-time PCR (qRT-PCR) analysis found treZ to be 16-fold higher in cultures containing ethanol. These results are in line with our microarray analysis, indicating that ethanol stimulates trehalose gene expression in both planktonic cultures and colony biofilms, as others have shown similarities in gene expression patterns in the two growth states. In contrast, an in-frame ΔalgU mutant had no significant difference in treZ expression between cells grown with and without ethanol (Fig. 2A). Like that of treZ, the expression of another AlgU-regulated gene, osmC, was higher (8-fold) upon growth with ethanol, and as expected, the differential expression was dependent on AlgU (Fig. 2B). The ΔalgU mutant also did not show an increase in trehalose upon growth with ethanol (Fig. 2C), and its defect could be complemented by restoring algU to the native locus (Fig. 2C). While the sigma factor AlgU was required for the induction of treZ transcripts and trehalose production, a mutant lacking another sigma factor, RpoS, which has been shown to regulate trehalose levels in E. coli (55), did not differ from the wild type in its response to ethanol (see Fig. S4 in the supplemental material).
FIG 2.
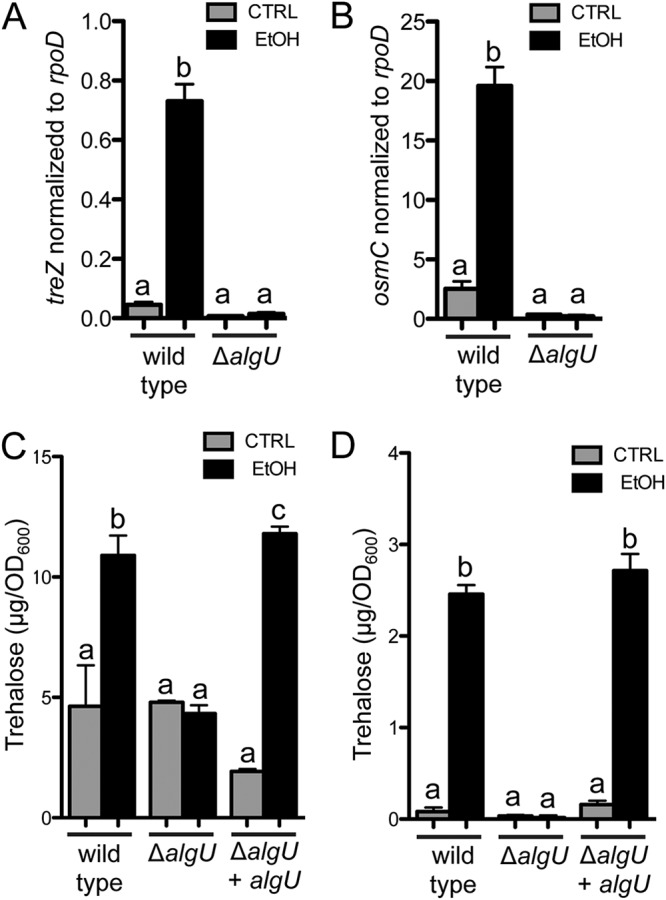
Analysis of the effects of ethanol on treZ and osmC transcript levels and intracellular trehalose in P. aeruginosa wild-type and ΔalgU strains. (A and B) treZ (A) and osmC (B) transcript levels relative to rpoD after growth in the absence and presence of 1% ethanol in buffered LB for 16 h. a-b, P < 0.0001 (two-way ANOVA and Tukey’s multiple-comparison test). (C and D) Trehalose levels in wild type, ΔalgU, and ΔalgU plus algU strains at the native locus in buffered LB (C) and M63 medium (D) with and without 1% ethanol for 16 h. (C) a-b, P < 0.002; a-c, P < 0.0009; b-c, not significant (NS). (D) a-b, P < 0.0001. (P values based on two-way ANOVA and Tukey’s multiple-comparison test.) The data are representative of the results of at least three independent experiments, each with at least three biological replicates. The error bars indicate standard deviations.
AlgU-dependent induction of trehalose in response to ethanol is independent of MucA cleavage and KinB regulation.
AlgU activity is repressed by the anti-sigma factor MucA, and proteolysis of MucA is a well-characterized means by which AlgU and its orthologs are activated (24, 53, 54, 56–60). MucA is bound by the periplasmic protein MucB, which inhibits MucA cleavage (61–63). Several stimuli, including high concentrations of ethanol (>3%) (26, 27), can lead to periplasmic stress and RpoE activation by anti-sigma factor degradation in other species. Several lines of evidence suggest that MucA cleavage was not the mechanism by which ethanol stimulated levels of treZ mRNA and trehalose. First, we found that ethanol increased trehalose to similar extents in wild-type and mucB mutant cells in a strain PA14 background (Fig. 3A); the mucB mutant has decreased stability of MucA, and the strain is mucoid, which indicates higher AlgU activation of genes involved in alginate biosynthesis. Second, ethanol stimulated trehalose levels in P. aeruginosa strain PAO1 in which mucA contained the mucA22 mutation (Fig. 3B). The truncated MucA22 mutant, a variant frequently found among P. aeruginosa clinical isolates, is no longer regulated by proteolysis and no longer represses AlgU (64, 65). Lastly, ethanol also stimulated trehalose in the alginate-overproducing mucoid cystic fibrosis isolate FRD1 (66), which also has the mucA22 allele (62, 67) (Fig. 3C). In FRD1, the stimulation of trehalose levels by ethanol required the presence of AlgU, as the isogenic nonmucoid algT/U::Tn501 (FRD440) derivative showed a large reduction in ethanol-stimulated trehalose (21) (Fig. 3C). These analyses showed that (i) ethanol induced responses similar to those in strain PA14 in the genetically distinct strains PAO1 and FRD1 and (ii) the increase in trehalose in response to ethanol does not require MucB or full-length MucA.
FIG 3.
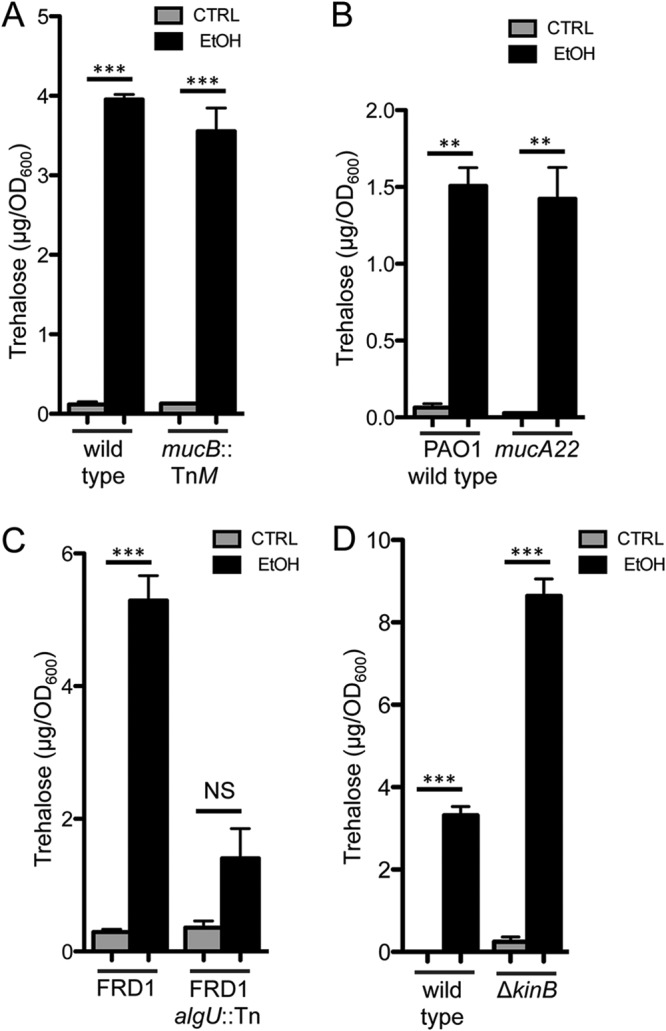
Trehalose levels in response to ethanol are independent of MucB, MucA cleavage, and KinB. Shown are trehalose levels of P. aeruginosa strain PA14 wild type and the validated mucB transposon mutant (A), P. aeruginosa strain PAO1 wild type and a mucA (MucA22) mutant (B), FRD1 and its isogenic algU::Tn derivative (C), and P. aeruginosa strain PA14 wild type and the ΔkinB mutant (D). Cultures were grown in M63 medium with and without 1% ethanol for 16 h. The data are representative of the results of at least 2 experiments, each with 3 biological replicates. The statistics are based on two-way ANOVA and Tukey’s multiple-comparison test. ***, P < 0.0001; **, P ≤ 0.0002. The error bars indicate standard deviations.
kinB loss of function has been associated with increased AlgU activity (53, 54) due to increased activity of the AlgB transcription factor, which stimulates AlgU expression; KinB phosphatase activity normally represses AlgB (68). Thus, we determined if KinB was required for the difference in trehalose levels in cells grown with or without ethanol. While the kinB mutant consistently had higher levels of trehalose under control and ethanol conditions than the wild type, KinB was not required for the effects of ethanol on trehalose levels (Fig. 3D). Together, these data support our model in which AlgU regulates trehalose through a mechanism that is not dependent on KinB.
Ethanol stimulation of trehalose requires SpoT-generated ppGpp.
In E. coli, the activity of RpoE, an AlgU homolog, is influenced by (p)ppGpp (34, 38, 39, 69, 70); (p)ppGpp effects on P. aeruginosa AlgU have not yet been reported. (p)ppGpp can be synthesized by either of two enzymes, RelA or SpoT (71–73). SpoT also has (p)ppGpp-degrading activity, and because very high levels of (p)ppGpp are toxic, mutants lacking spoT are viable only in the absence of RelA activity (74, 75). Thus, we determined if ethanol stimulation of trehalose levels was altered in either the ΔrelA or ΔrelA ΔspoT strain. We found that while the ΔrelA strain was like the wild type, ethanol did not influence trehalose levels in the ΔrelA ΔspoT double mutant, suggesting that SpoT was required for trehalose accumulation in ethanol-grown cells (Fig. 4A). Complementation of the ΔrelA ΔspoT double mutant with spoT restored trehalose levels to those in the wild type in the presence of ethanol (see Fig. S6 in the supplemental material). The induction of treZ and osmC was also dependent on SpoT (Fig. 4C and D).
FIG 4.
dksA and spoT are required for trehalose accumulation and treZ and osmC expression, and spoT is necessary for increased (p)ppGpp in response to ethanol. (A and B) Trehalose levels of PA14 wild type, ΔrelA, and ΔrelA ΔspoT (A) and PA14 wild-type, ΔdksA, and ΔdksA plus dksA (B) strains in M63 medium with and without 1% ethanol for 16 h. (C and D) treZ (C) and osmC (D) transcripts normalized to rpoD in PA14 wild-type, ΔdksA, and ΔrelA ΔspoT strains in buffered LB at 16 h with and without 1% ethanol. The data are representative of the results of at least 3 independent experiments with at least 3 biological replicates each. (E) (p)ppGpp quantification (relative fluorescence units [RFU], 470/380 nm/OD600 unit) of PA14 wild-type, ΔrelA, and ΔrelA ΔspoT strains in M63 medium with and without 1% ethanol for 16 h. The data are representative of the results of at least 2 independent experiments. The statistics are based on two-way ANOVA and Tukey’s multiple-comparison test. (A) a-b, P = 0.0179; a-c and d-e, P = 0.0001; a-d, a-e, b-d, b-e, c-d, and c-e, P < 0.0001; b-c, NS. (B) a-b, P < 0.0001. (C) a-b and b-c, P < 0.0001; a-c, P ≤ 0.05. (D) a-b, P < 0.0001. (E) a-b, P < 0.0001. The error bars indicate standard deviations.
To determine if 1% ethanol altered (p)ppGpp levels in the PA14 wild type, its levels were measured. We found that cells grown with ethanol had 3.45-fold higher levels of (p)ppGpp. The increase in (p)ppGpp was similar to that of the wild type in the relA mutant, but the ΔrelA ΔspoT strain did not show an increase in (p)ppGpp in response to ethanol, indicating that ethanol stimulates (p)ppGpp production via SpoT (Fig. 4E).
Ethanol stimulation of treZ and trehalose levels requires DksA.
The (p)ppGpp signal influences the activity of RNA polymerase-sigma factor complexes through DksA, an RNA polymerase binding protein. We found that a ΔdksA mutant no longer showed ethanol-induced trehalose accumulation and that the phenotype of the ΔdksA mutant was complemented by restoring dksA at the native locus (Fig. 4B). Consistent with the trehalose measurements, ethanol-induced increases in treZ and osmC expression were greatly reduced in the ΔrelA ΔspoT and ΔdksA strains (Fig. 4C and D). Like those of the wild type, the growth kinetics of the ΔdksA mutant and the ΔrelA ΔspoT mutant were not reduced by 1% ethanol, though the ΔdksA mutant grew more slowly than the wild type under control conditions, consistent with published reports (76, 77) (see Fig. S5 in the supplemental material).
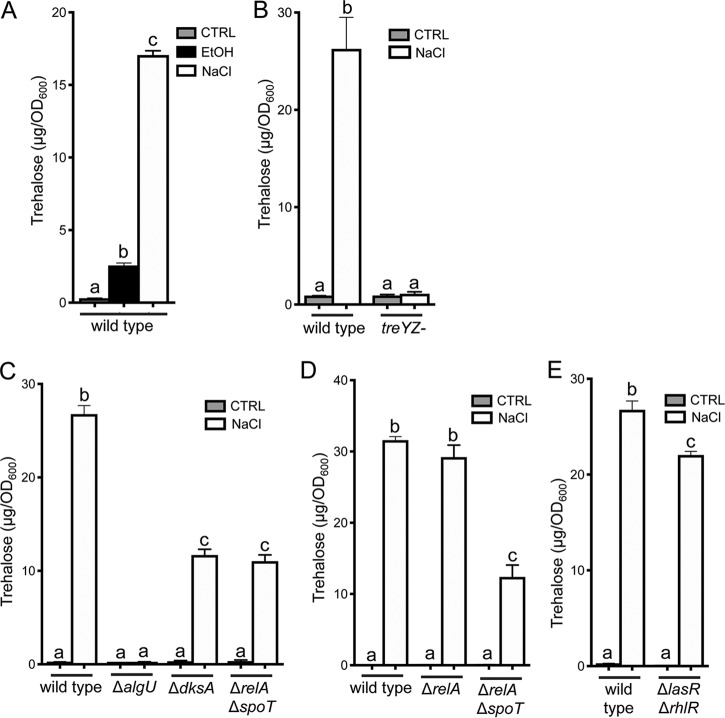
AlgU is required for, and SpoT and DksA contribute to, the induction of trehalose by high NaCl.
In P. aeruginosa and other species, trehalose is induced by high NaCl levels (46, 78). Ausubel and colleagues (43) found that elevated trehalose levels under high-NaCl conditions required TreYZ (43), and we confirmed these results (Fig. 5A and B). Furthermore, we found that trehalose induction in response to high NaCl was absent in the ΔalgU mutant (Fig. 5C), as it was for ethanol (Fig. 1C and D). While (p)ppGpp and DksA were necessary for induction of trehalose in response to ethanol, the ΔdksA and ΔrelA ΔspoT strains still showed significant induction of trehalose in response to NaCl (Fig. 5C). The level of induction, however, was significantly lower in the ΔdksA and ΔrelA ΔspoT strains than in the wild type and the ΔrelA strain, suggesting that SpoT-dependent (p)ppGpp and the (p)ppGpp-responsive RNA polymerase binding protein DksA contributed to the response in cells from post-exponential-phase cultures (Fig. 5C and D).
FIG 5.
Trehalose accumulation in response to NaCl requires TreYZ and AlgU. (A) NaCl (500 mM) stimulates trehalose accumulation. (B) NaCl-stimulated accumulation of trehalose is dependent on the TreYZ pathway. (C) NaCl-stimulated trehalose accumulation requires algU, and dksA and spoT contribute. (D) NaCl-stimulated trehalose accumulation is independent of relA. (E) Trehalose accumulation in NaCl in the ΔlasR ΔrhlR strain is similar to that in the wild type. Cultures were grown for 16 h in M63 medium with 1% ethanol or 500 mM NaCl as indicated. The data are representative of the results of two or more experiments, each with at least 3 biological replicates. The statistics are based on one-way ANOVA (A) or two-way ANOVA (B, C, and D) and Tukey’s multiple-comparison test. (A) a-b, P = 0.0005; a-c and b-c, P < 0.0001. (B) a-b, P < 0.0001. (C) a-b, a-c, a-d, and b-c, P < 0.0001. (D) a-b, a-c, a-d, and b-c, P < 0.0001. (E). a-b and a-c, P < 0.0001; b-c, P = 0.0007. The error bars indicate standard deviations.
Quorum-sensing master regulators are necessary for the ethanol induction of trehalose.
Schuster et al. (40, 41) found that treZ and other genes involved in trehalose biosynthesis and osmC were at lower levels in a PAO1 ΔlasR ΔrhlR mutant than in the wild type. They reported that treZ and osmC fell within a subset of QS-controlled genes that were induced later in growth in batch culture than other QS-controlled genes (40, 41). We found that the increase in levels of treZ and osmC (Fig. 6A and B) or trehalose (Fig. 6C) in ethanol-grown cells did not occur in the ΔlasR ΔrhlR strain and that the lack of response did not appear to be due to ethanol effects on growth in the strain (see Fig. S5). Analysis of trehalose levels in lasR and rhlR single mutants was also performed. We found that lasR was necessary for the accumulation of trehalose in response to ethanol and that the defect in the ΔlasR mutant was complemented by providing lasR back at the native locus (see Fig. S7 in the supplemental material). The ΔrhlR mutant phenotype was consistent within an experiment but variable across experiments (discussed further below) (see Fig. S7). Since lasR or both AHL-responsive transcription factors were necessary for the stimulation of trehalose in cells grown with ethanol, we determined if ethanol enhanced AHL-mediated quorum sensing, thereby inducing trehalose levels. To do so, we monitored the expression of rhlI, a quorum-sensing-controlled gene regulated by LasR and RhlR, using an rhlI-lacZ promoter fusion (79). PA14 wild type and the ΔlasR ΔrhlR strains were grown with or without ethanol. As expected, levels of β-galactosidase (β-Gal) activity were much lower in the ΔlasR ΔrhlR mutant than in the wild type, and we found no significant difference in rhlI promoter activity in ethanol-grown cells compared to cells from control conditions (Fig. 6D). The transcriptomics analysis of P. aeruginosa grown with or without ethanol as described above did not find evidence for ethanol affecting AHL-mediated quorum sensing broadly (see Table S3A and B). Additionally, while AHL quorum sensing was required for trehalose accumulation in response to ethanol, it was not necessary for trehalose accumulation in response to high NaCl (Fig. 5E); there was, however, a significant reduction in trehalose levels in the ΔlasR ΔrhlR strain compared to the wild type in NaCl, suggesting that this mechanism played a role.
FIG 6.
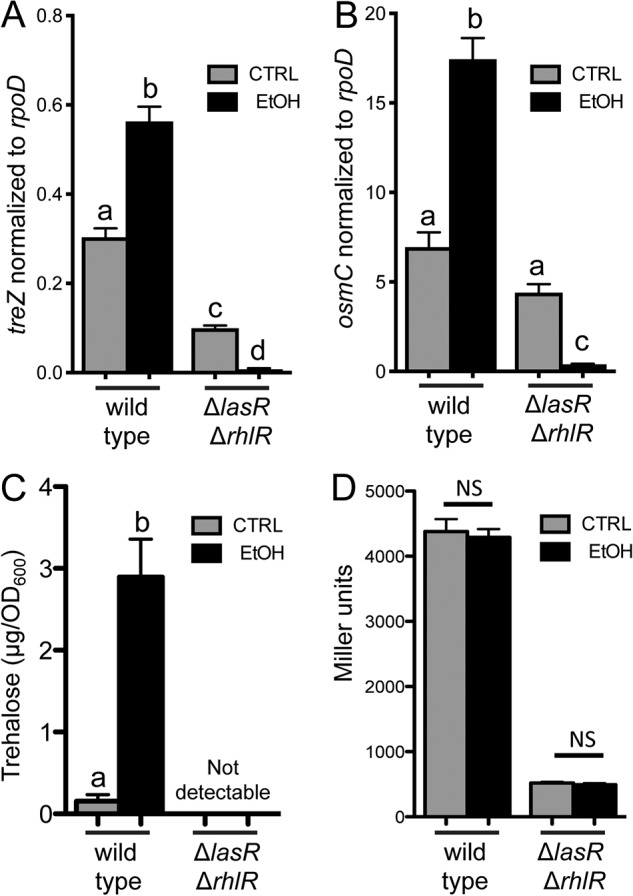
AHL quorum-sensing regulation is required for trehalose accumulation and increased treZ and osmC transcripts in response to ethanol. (A and B) Expression of treZ (A) and osmC (B) genes normalized to rpoD in PA14 wild-type and ΔlasR ΔrhlR strains in buffered LB with and without 1% ethanol for 16 h. (C) Trehalose quantification of PA14 wild-type and ΔlasR ΔrhlR cells grown planktonically in M63 medium with and without 1% ethanol for 16 h. (D) β-Galactosidase assay of rhlI-lacZ promoter activity in PA14 wild-type and ΔlasR ΔrhlR cells grown planktonically in M63 medium with and without 1% ethanol for 16 h. The data are representative of the results of at least 2 independent experiments, each with 3 or more biological replicates. The statistics are based on two-way ANOVA and Tukey’s multiple-comparison test. (A) a-b, a-d, b-c, and b-d, P < 0.0001; a-c, P = 0.0004; c-d, P = 0.0395. (B) a-b and b-c, P < 0.0001; a-c, P < 0.02. (C) a-b, P = 0.0002. The error bars indicate standard deviations.
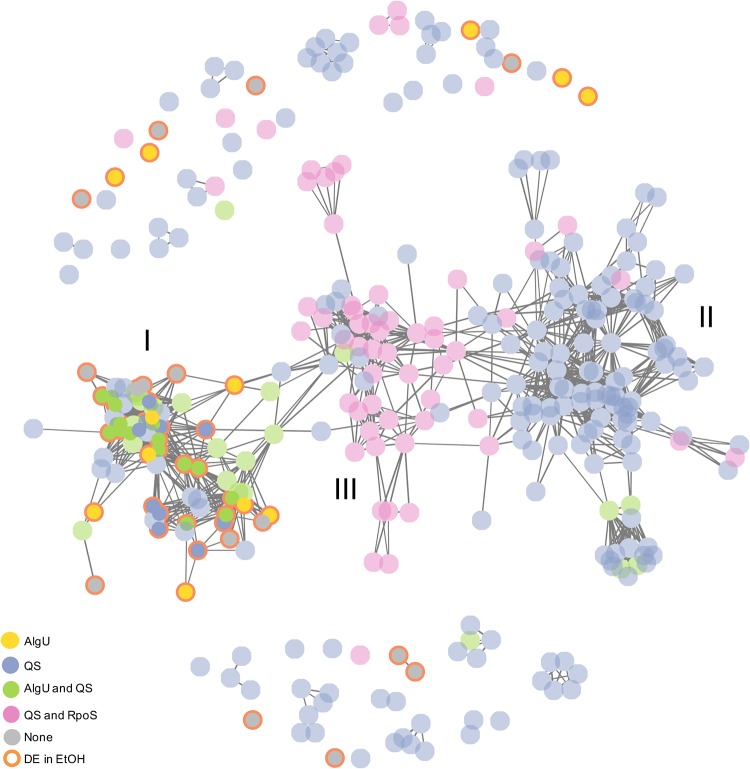
Ethanol-responsive genes comprise a distinct cluster within a structured network of QS-controlled genes also regulated by AlgU.
Together, our data present a complex scheme in which global regulators, AlgU and transcription factors involved in AHL-mediated quorum sensing, control the trehalose-biosynthetic genes treYZ and osmC and levels of trehalose in cells grown with ethanol. Our findings support previous reports that separately found treZ and osmC genes to be among the genes controlled by AlgU (51, 52) and AHL-mediated quorum sensing (40, 41). To test the hypothesis that a subset of genes are members of both the AlgU and QS regulons and that ethanol specifically altered the expression of this subset of genes, we used eADAGE, a machine learning algorithm used to generate a model for P. aeruginosa expression patterns from 1,051 publicly available transcriptome samples (42, 80, 81). The eADAGE model learned 600 expression signatures, and within each signature, genes had different weights. Similarities in weights across signatures for genes indicate a correlation in expression levels across the large P. aeruginosa transcriptomics data set. Pairwise Pearson correlation coefficients of the genes in the eADAGE model can be visualized as edge weights in a network where nodes are genes (eADAGE [80]).
In the network shown in Fig. 7, we present the relationships in expression patterns for (i) genes that were found to be differentially expressed in response to ethanol (nodes with orange borders) and (ii) the set of genes differentially expressed by more than 5-fold in a ΔlasR ΔrhlR strain compared to the wild type, which included treZ and osmC, as reported by Schuster et al. (40) (blue nodes) (a gene list is shown in Table S3D). Using a data set that characterized the AlgU regulon by comparing a ΔalgU mutant to the wild type under AlgU-inducing heat shock conditions (51), we identified genes in the two data sets mentioned above that were regulated by AlgU (yellow nodes) (see Table S3C for a gene list) or that were altered in both the ΔlasR ΔrhlR and ΔalgU strains (green nodes).
FIG 7.
Ethanol-responsive genes are enriched for those with overlapping AlgU and QS control. eADAGE gene-gene network analysis shows ethanol-responsive genes and the QS regulon, with overlapping AlgU genes indicated. (I) Enrichment region for genes coordinately regulated by AlgU and QS (green) and ethanol-responsive genes (orange outline). (II) Enrichment region for “canonical” quorum-sensing genes, including lasI, lasB, rhlR, rhlI, and rhlA. (III) Enrichment region for QS genes within the RpoS regulon.
The majority of genes included in the network were connected by edges, revealing strongly correlated expression patterns across the large data compendium comprised of the results of experiments performed by different laboratories with different strains and under different conditions over more than a decade (42, 80, 81). The connected genes fell into three major clusters (I, II, and III). The majority (80%) of the QS-controlled genes that were also AlgU controlled (Fig. 7, green nodes) (52) were found in cluster I. Ethanol-responsive transcripts (orange borders), including treZ and osmC, were exclusively localized to cluster I. Statistical analysis found that genes differentially expressed in response to ethanol (see Table S3A) represented 12.2% of AlgU-controlled genes (51) and 8.6% of QS-controlled gene sets as defined above (see Table S3 for gene lists) but comprised 44% of the genes present in both regulons. The enrichment of the intersection of AlgU- and QS-controlled genes over the sets of either all AlgU- or all QS-regulated genes was significant (P = 0.001 and P = 0.00002, respectively). Visualization of a cluster of ethanol-responsive genes within the gene-gene network comprised of the complete AlgU regulon (51) is shown in Fig. S8 in the supplemental material.
Cluster II genes contained many genes known to be regulated mainly by LasR or RhlR and their cognate signals (40, 41). Examples of genes in cluster II are lasI, lasB, rhlR, rhlI, and rhlA. The lack of any of the ethanol-responsive genes in cluster II is consistent with our finding that ethanol did not alter expression of rhlI (Fig. 6D) and supports our model in which ethanol did not broadly induce the entire QS regulon.
Cluster III contained genes from the QS-controlled gene set that were previously described by Schuster and Greenberg (41) as also being differentially expressed in an rpoS mutant (Fig. 7, pink nodes) (see Table S3E for a gene list). Of the 56 genes in cluster III, 47 (41) and 29 (51) genes were differentially expressed in separate published studies describing the RpoS regulon. Ethanol-responsive genes were not among the genes in cluster III, supporting the above-mentioned data showing that ethanol-induced trehalose levels were not dependent on rpoS (see Fig. S4). Specific enrichment in AlgU- and QS-coregulated genes among the genes upregulated in response to ethanol is consistent with ethanol activating only a subset of the AlgU- and AHL-controlled regulons.
Ethanol induces trehalose after entry into post-exponential phase.
Increased trehalose levels and osmC and treZ transcripts in cultures with ethanol was dependent on both SpoT-synthesized (p)ppGpp and AHL-mediated quorum sensing; signals associated with growth restriction, often due to nutrient limitation; and high cell density, respectively. Analysis of trehalose levels in control and ethanol-containing cultures found that ethanol affected trehalose levels only in cells from post-exponential-phase cultures but not in cells from exponential-phase cultures. This relationship was observed in both M63 medium (Fig. 8A) and buffered LB medium (1.96 μg/optical density at 600 nm [OD600] unit in control cultures versus 2.81 μg/OD600 unit in ethanol-containing cultures; P = 0.1339). Expression of treZ was also not affected by ethanol in cells from exponential-phase cultures but was affected in cells from the same cultures collected after entry into post-exponential phase (Fig. 8B). Together, these data support a model in which the induction of trehalose in response to ethanol by AlgU requires other signals from quorum-sensing- and growth-restriction-associated pathways.
FIG 8.
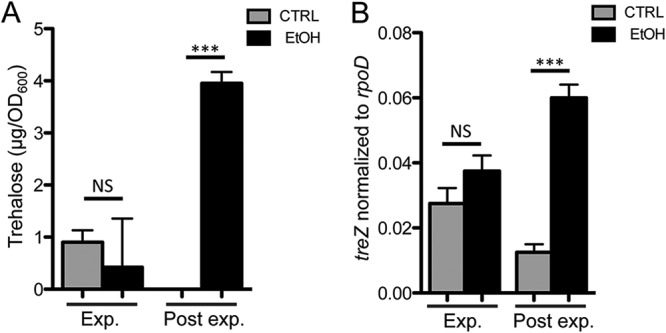
Ethanol stimulates treZ gene expression and trehalose levels in cells from post-exponential-phase (Post exp.) but not exponential-phase (Exp.) cultures. (A) Trehalose quantification of cells grown in M63 medium with and without 1% ethanol for 6 h to an OD600 of ∼1.0 (Exp.) or for 16 h (Post exp.). (B) treZ gene expression normalized to rpoD in cultures grown in buffered LB with and without 1% ethanol to the same densities as for panel A. The data are representative of the results of 3 experiments, each with 2 to 4 biological replicates. The statistics are based on two-way ANOVA and Tukey’s multiple-comparison test. ***, P ≤ 0.0008. The error bars indicate standard deviations.
DISCUSSION
The data presented above led us to propose a model, based largely on genetic analyses, in which ethanol activates AlgU through stimulation of (p)ppGpp, synthesized by SpoT, and activation of DksA-dependent transcription. Chromatin immunoprecipitation experiments have shown that AlgU binds to the promoter for the treYZ-containing operon (51). AHL-mediated quorum sensing through LasR and/or RhlR was required for AlgU-dependent activation of treZ and osmC and increased levels of trehalose, and thus ethanol induction of trehalose was observed in cultures only after AHL-mediated QS was induced. Our data do not indicate that ethanol led to a global increase in expression of the QS regulon. Based on these data, we propose that even though ethanol was present over the course of growth, increased trehalose biosynthesis in response to ethanol occurs only when cells have sensed a quorum and when (p)ppGpp synthesis can be stimulated, perhaps because of concomitant nutrient limitation signals, which are known to activate SpoT (Fig. 9). These data highlight a nuanced response to a microbially produced molecule, ethanol, in P. aeruginosa, and this underscores how microbe-microbe interactions may change with shifts in physiological states and extracellular signal concentrations.
FIG 9.
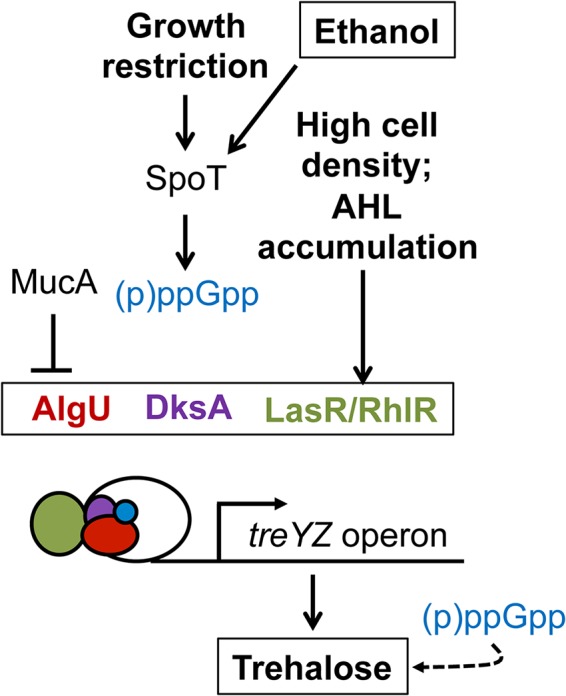
Model describing ethanol stimulation of the AlgU regulon and trehalose production. Our data support a model in which cell density and growth signals determine the cell’s response to ethanol. The colors of the text are reflected in the schematic of the transcriptional-regulatory complex.
It is not surprising that we saw variation in the levels of trehalose induced by ethanol given that multiple dynamic systems are involved in its regulation. The trehalose levels in cultures with ethanol were statistically significantly higher than those in control cultures in 23 independent experiments performed on different days, with each containing three biological replicates inoculated from three different overnight cultures. If each experiment was treated as an independent data point, the average increase in trehalose levels in response to ethanol was 3.5 μg/OD600 unit with a P value of <0.0001. In the absence of ethanol, trehalose levels ranged from undetectable to 0.5 μg/OD600 unit, and in the presence of 1% ethanol, trehalose values ranged from 2 to 6.4 μg/OD600 unit.
The model for AlgU activation by (p)ppGpp and DksA represents a new mechanism by which the important sigma factor AlgU may be activated in P. aeruginosa. (p)ppGpp-dependent activation of E. coli RpoE, an AlgU ortholog, has been reported previously (34) and has been associated with the growth phase (82), but activation by noninhibitory concentrations of ethanol has not been reported. Our data showed that ethanol, at a 1% concentration, did not activate AlgU through effects on the MucA or MucB protein (Fig. 3), which are targeted for cleavage in response to periplasmic or envelope stress. In the presence of 0.5 M NaCl, however, a stimulus that is expected to induce periplasmic stress, DksA- and SpoT-independent stimulation of trehalose was observed, presumably because some AlgU activation occurred through MucA cleavage, but DksA and SpoT did contribute to the strength of the response, suggesting that these mechanisms can work together.
The intersection of QS and AlgU regulation is interesting. Future experiments will also determine if LasR and/or RhlR directly interacts with the promoter upstream of the treYZ-containing operon and if that interaction occurs only when AlgU and DksA are complexed with RNA polymerase. Evidence for direct activation of osmC and treYZ expression by LasR comes from the finding that overexpression of lasR but not rhlR was sufficient to induce their expression. Consistent with this, the ΔlasR strain was defective in the induction of trehalose. While we observed little variability between replicates within each experiment, we did observe variability in the importance of ΔrhlR across experiments, suggesting that an unidentified variable influenced the need for RhlR for treYZ induction. Thus, we propose that either transcription factor can influence the expression of treYZ, perhaps with different kinetics, as has been shown for other genes (83). In separate studies with ΔdksA and ΔrelA ΔspoT mutants in strain PAO1, DksA and (p)ppGpp have independently been associated with both the positive and negative regulation of genes that are differentially expressed in a ΔlasR ΔrhlR mutant (77, 84–88) so there may be a complex relationship between these signaling pathways.
The biological role of AlgU-induced trehalose in P. aeruginosa in response to stresses or in microbial communities is not yet known. AlgU has been implicated in both the positive and negative regulation of genes involved in oxidative and osmotic stress responses in other pseudomonads (89–92). For example, in different pathovars of P. syringae, AlgU regulates oxidative- and osmotic-stress response genes transcriptionally in response to osmotic stress (91) and contributes to plant disease independently of alginate (92). In Pseudomonas fluorescens, an algU mutant was significantly more sensitive to osmotic stress than the wild type (89). P. fluorescens AlgU appears to be necessary for desiccation stress tolerance but dispensable for tolerance of 3% hydrogen peroxide, 1.9% paraquat, 5% sodium hypochlorite, heat shock, pH extremes, and the reducing agent dithiothreitol (89). In P. aeruginosa, AlgU has been described as having a negative role in oxidative-stress resistance and can be protective against host innate immune factors through its regulation of alginate (24, 25). We did not observe a protective benefit of growth with 1% ethanol in oxidative stress, osmotic stress, and dessication assays under the growth conditions used in these studies (data not shown). Trehalose has been reported to protect against osmotic, oxidative, heat, and cold stress by stabilizing proteins and reducing the formation of denatured protein aggregates (93–97) and to act as a carbon reserve (93, 98), and recent studies have found that trehalose can stabilize outer membrane vesicles (99). Trehalose can accumulate in the cytoplasm and periplasm and be secreted, making it an interesting molecule to consider in the context of microbe-microbe interactions. P. syringae survival as an epiphyte and P. aeruginosa plant pathogenesis both require the ability to make trehalose (43, 100). Interestingly, exogenous trehalose and wild-type-derived trehalose in coculture rescued the attenuated trehalose mutant phenotype in P. aeruginosa Arabidopsis pathogenicity in planta, with the requirement for trehalose being independent of osmoprotection (43). Pseudomonads can accumulate a variety of osmoprotectants in addition to trehalose, including betaine, ectoine, and N-acetylglutaminylglutamine amide (NAGGN) (100). This redundancy may contribute to the fact that trehalose mutants are not more sensitive to tested stresses in laboratory assays (43).
In addition to treZ and other trehalose metabolism genes, cluster I included 38 genes found within the genome island that spans from PA2134 to PA2192 (PA14_36980 to PA14_36345) (54, 101, 102). Sixteen of the 54 genes (∼30%) differentially expressed in response to ethanol were in this chromosomal region. Many of the genes in this chromosomal region that are differentially expressed in ethanol could participate in survival of stresses likely to be present in mixed-species communities formed with ethanol-producing microbes. For example, glgE is involved in the metabolism of glycogen, a carbon and energy storage molecule that accumulates when carbon is in excess relative to other growth-limiting nutrients (103). Other genes within the genome island encode putative ion transporters, double-strand break repair enzymes, and two catalases. Other ethanol-induced genes within cluster I included osmC, sprP, and pfpI. OsmC can be protective against oxidative stress caused by exposure to elevated osmolarity and hyperoxides through an unknown mechanism (104). SprP is a subtilase protease (105), and pfpI codes for a protease that plays a role in DNA protection under nonstress conditions and in the presence of hydrogen peroxide (106). Mutation of either sprR or pfpI in P. aeruginosa has pleiotropic effects (105, 106).
The next important question involves the mechanism by which ethanol stimulates (p)ppGpp. Others have shown that (p)ppGpp levels increase in response to higher concentrations of ethanol and that in E. coli, the addition of ethanol mimics amino acid starvation (107). They speculated that ethanol (and other short-chain alcohols) may interfere with amino acid uptake (107). Ethanol may also directly impact ribosome activity (108) or other pathways through effects on cell membranes, and the fact that responses vary based on the growth phase provides a useful tool to understand how ethanol and ethanol-producing microbes influence other bacteria.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Bacteria were maintained on 1.5% agar LB plates (109). Where stated, ethanol (200 proof) was added to the medium (liquid or molten agar) to a final concentration of 1% unless otherwise stated. NaCl to a final concentration of 500 mM was added to liquid media as indicated. Mutants from the PA14 nonredundant (NR) library were grown on LB with 60 μg/ml gentamicin (49). When strains from the NR library were used, the location of the transposon insertion was confirmed using site-specific primers. The primers are listed in Table S2 in the supplemental material. Where stated, LB medium was buffered to pH 8 with 100 mM HEPES buffer (referred to as buffered LB). M63 medium contained 0.2% glucose and 2% Casamino Acids (110). When ethanol was supplied as a sole carbon source, glucose and amino acids were omitted. Planktonic cultures were grown at 37°C on a roller drum.
For the growth curves shown in Fig. S1, overnight cultures were diluted in 5 ml fresh buffered LB medium in 18- by 150-mm borosilicate glass tubes with or without 1% ethanol to an OD600 of ∼0.05 and incubated at 37°C on a roller drum. Similar culture vessels and volumes were used in all assays unless otherwise specified. OD600 measurements were taken on a Genesys 6 spectrophotometer. For the data in Fig. S5, overnight cultures were diluted in fresh LB medium buffered to pH 8 with HEPES with or without 1% ethanol to an OD600 of ∼0.05, and 150 μl of the cell suspension was pipetted into 96-well plates. The plates were grown at 37°C with continuous shaking at ∼150 rpm. OD600 measurements were taken at 16 h of incubation using a microplate reader.
Construction of in-frame deletions, complementation, and plasmids.
Construction of plasmids, including in-frame deletion and complementation constructs, was completed using yeast cloning techniques in Saccharomyces cerevisiae as previously described (111) unless otherwise stated. The primers used for plasmid construction are listed in Table S2 in the supplemental material. In-frame deletion and single-copy complementation constructs were made using the allelic-replacement vector pMQ30 (46). Deletions of relA and spoT were introduced in the PA14 strain using the pEX18Gm suicide vector to create unmarked deletion mutants (112), as previously described (113). Promoter fusion constructs were made using a modified pMQ30 vector with a lacZ-GFP fusion integrating at the neutral att site on the chromosome. The rhlI promoter region was amplified from PA14 genomic DNA (gDNA) using Phusion high-fidelity DNA polymerase with primer tails homologous to the modified pMQ30 ATT KI vector containing the lacZ-GFP reporters. The 195-bp upstream promoter region includes an RhlR-binding site as annotated at the Pseudomonas Genome Database (121) at positions −192 to +3. All the plasmids assembled using yeast cloning were purified from yeast using Zymoprep Yeast Plasmid Miniprep II according to the manufacturer's protocol and transformed into electrocompetent E. coli strain S17 by electroporation. The spoT complementation construct was made by linearizing the pMQ30 vector using EcoRI. The linearized plasmid was treated with shrimp alkaline phosphatase purchased from New England Biolabs and cleaned using a Qiagen PCR purification kit (Qiagen, Hilden, Germany). spoT fragment 1 was amplified from P. aeruginosa PA14 genomic DNA for a 1,369-bp PCR product. spoT fragment 2 was amplified from PA14 genomic DNA for a 1,468-bp PCR product. spoT fragment 3 was amplified from PA14 genomic DNA for a 1,268-bp PCR product. All amplified DNA was treated with DpnI to remove any remaining template and then cleaned using a Qiagen PCR purification kit. The vector and spoT fragments were assembled using NEBuilder HiFi DNA assembly master mix and incubated at 60°C for 1 h. The DNA was then transformed into high-efficiency DH5α chemically competent cells. The plasmid was isolated using a Zyppy plasmid miniprep kit (Zymo Research, Irvine, CA) and then screened by digestion with NruI. The plasmid was confirmed by sequencing. Plasmids were introduced into P. aeruginosa by conjugation, and recombinants were obtained using sucrose counterselection and genotype screening by PCR.
Microarray analysis.
Cultures of P. aeruginosa PA14 wild type were grown overnight in LB at 37°C on a roller drum, and 5 μl of the overnight culture was spotted onto tryptone plates (1% tryptone, 1.5% agar) (114) with or without 1% ethanol. The plates were incubated at 37°C for 16 h. Colonies were scraped up from the plates for RNA isolation using a Qiagen RNeasy Mini kit. The samples were DNase treated using an Invitrogen Turbo DNA-Free kit. As previously described (115), cDNAs labeled with biotin-ddUTP (Enzo Bio-Array terminal labeling kit; Affymetrix) were hybridized to Pseudomonas GeneChips using a GeneChip fluidics station 450 (Affymetrix) according to the manufacturer’s instructions. The GeneChips were scanned in the Dartmouth Genomics and Microarray Laboratory using a GeneChip Scanner 3000 7G (Affymetrix), and the BioConductor Affy library was used to read CEL file data. The data were normalized with RMA in BioConductor (116).
eADAGE analysis.
Genes upregulated 2-fold or more were analyzed within the context of their expression patterns in a compendium of 1,051 publicly available microarrays from the Gene Expression Omnibus as determined by a machine learning model, eADAGE (42). Pearson correlations of greater than 0.5 between the learned parameters corresponding to each gene were visualized by edge weights in the resultant network (80). Genes differentially expressed between the wild type and a ΔlasR ΔrhlR mutant strain sampled at different time points over the course of growth are referred to as the QS-controlled regulon (40) (see Table S3D). The QS-controlled genes are presented as a network, plotted using the Fruchterman-Reingold force-directed algorithm, in which correlations in gene expression are indicated with the presence of edges and genes with shorter edges are more strongly correlated in expression pattern. The network was generated in R using the “network” (117, 118), “GGally” (119), and “ggplot2” (120) packages. The genes within the QS-controlled gene set that were also differentially expressed upon deletion of algU (51) are indicated as green nodes, and genes within the QS-controlled gene set that were also differentially expressed upon deletion of rpoS (41) (see Table S3E) are indicated as pink nodes. QS-controlled genes that were not differentially expressed upon deletion of AlgU or RpoS are presented as blue nodes. The complete gene list for each data set and accompanying R code are available in the supplemental material. If necessary, PA14 gene numbers gene were converted to PAO1 ortholog gene numbers, and PAO1 gene numbers were converted to gene names, using P. aeruginosa PA14 109 orthologs and P. aeruginosa PAO1 107 annotations from the Pseudomonas Genome Database (121).
Quantitative-PCR analysis of transcripts.
For quantitative real-time PCR experiments, cultures of the indicated strains of P. aeruginosa were grown for 16 h in 5 ml of buffered LB at 37°C on a roller drum. RNA was isolated from planktonic cultures using a Qiagen RNeasy Mini kit. The samples were DNase treated using an Invitrogen Turbo DNA-Free kit. cDNA was synthesized with a RevertAid H-minus first-strand synthesis kit using the GC-rich protocol at the following temperatures: 25°C for 5 min, 50°C for 60 min, and 70°C for 5 min. The synthesized cDNA was diluted 1:5 in molecular-grade water and stored at −20°C. Quantitative-PCR expression analysis was performed using an Applied Biosystems 7500 real-time PCR system with Bio-Rad SsoFast Evagreen Supermix and the primers listed in Table S2. A cycling regimen of 95°C for 30 s, 39 cycles of 95°C for 10 s and 60°C for 5 s, and a final 65°C for 3 s was used. Experimental transcripts were normalized to the housekeeping gene rpoD.
Measurement of trehalose in cells.
Trehalose was quantified from whole-cell lysates as described previously (122, 123) with slight modifications. Briefly, bacterial cultures were grown for 16 h in LB or M63 medium with or without 1% ethanol as indicated in 18-mm borosilicate culture tubes at 37°C on a roller drum. The cultures were inoculated from strains grown on LB plates. A 250-μl volume of culture was concentrated to an OD600 of 8.0 in sterile water. Cell suspensions were boiled for 10 min to lyse the cells. The resulting lysate was centrifuged at 16,000 × g, and 100-μl aliquots of lysate were transferred to new tubes. One tube of lysate was treated with 1 μl of trehalase (Sigma-Aldrich) enzyme or vehicle control. Glucose in the lysate samples was quantified using a glucose oxidase kit (Sigma; catalog no. GAGO20). Trehalose concentrations were calculated based on a standard curve and subtraction of the basal glucose and are expressed relative to OD units.
(p)ppGpp measurements.
Cultures inoculated at an OD600 of 0.05 were grown for 16 h to an OD600 of ∼2.0 in 5 ml M63 medium with and without 1% ethanol in 18-mm culture tubes at 37°C with shaking at 250 rpm. About 2 ml of each cultures was pelleted at 10,000 × g for 5 min, and the (p)ppGpp was extracted as described previously (124). Briefly, the pellets were suspended in 200 μl of 10 mM Tris-HCl, pH 7.8, containing 1 mg/ml lysozyme and 15 mM magnesium acetate. The suspensions were vortexed for 3 s and subjected to two freeze-thaw cycles. Then, 15 μl of a 10% deoxycholate solution was added, and the suspensions were vortexed for 15 s. Finally, 200 μl of 10 mM Tris-HCl, pH 7.8, containing 15 mM magnesium acetate was added, and the samples were centrifuged at 12,000 × g for 10 min. The supernatants were used for (p)ppGpp quantification, which was performed using the chemosensor Bis-Zn2+-dipicolylamine (PyDPA) as previously described (125). The measurements of (p)ppGpp-PyDPA complex were carried out immediately after probe addition using fluorescence spectroscopy (excitation wavelength [Ex]/emission wavelength [Em] = 344/480 nm) in a Tecan Infinite M1000 plate reader. To account for the interference of other nucleotides, ΔrelA ΔspoT mutant extracts were used, and the absolute (p)ppGpp values were determined using a calibration curve with purified (p)ppGpp (TriLink Biotechnologies) spanning a linear range of 0.4 to 6 μM (p)ppGpp. For normalized (p)ppGpp measurements, fluorescence at Ex/Em values of 344/480 nm was normalized by the fluorescence at Ex/Em values of 344/380 nm (free nucleotides complex with PyDA) and by the OD600 of the washed cultures.
Measurement of β-galactosidase in reporter fusion strains.
Cells with an rhlI promoter fusion to lacZ-GFP genes integrated at the attB locus were grown in 5 ml of LB at 37°C. About 1 ml of overnight culture was pelleted, washed twice, and resuspended in phosphate-buffered saline (PBS). The washed cells were diluted to a starting OD600 of 0.05 in 5 ml of M63 medium with and without 1% EtOH. After 16 h on a roller drum at 37°C, β-Gal activity was measured as described by Miller (126).
Statistics.
Unless otherwise stated, data are based on three biological replicates with the mean and standard deviations calculated and are representative of the results of at least three independent experiments containing multiple replicates. Unless otherwise stated, means and standard deviations were calculated in Graph Pad Prism 8, and analyses were completed using a two-way analysis of variance (ANOVA) and Tukey’s multiple-comparison test, with P values indicated in the figure legends. Regulon enrichments were determined by hypergeometric tests using the “phyper” function in the “stats” package in R.
Accession number(s).
The data for our microarray analysis of the P. aeruginosa PA14 wild type in response to ethanol has been uploaded to the GEO repository (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE124852.
Supplementary Material
ACKNOWLEDGMENTS
The research reported in this article was supported by National Institutes of Health (NIH) grants R01 GM108492 to D.A.H., NIAID T32AI007519 to C.E.H. and D.L.M., NIGMS T32-GM008704 to G.D., and NHLBI T32HL134598 to M.E.C.; Burroughs Wellcome Fund and Canadian Institutes of Health Research funding to D.N.; and Canadian Institutes of Health Research and Cystic Fibrosis Canada fellowships to D.M. Microarray processing was carried out at Dartmouth Medical School in the Genomics Shared Resource Core, which was established by equipment grants from the NIH and NSF and is supported in part by a Cancer Center Core Grant (P30CA023108) from the National Cancer Institute. Support for the project was also provided by NIGMS P20GM113132 through the Molecular Interactions and Imaging Core (MIIC) and CF RDP STANTO19R0 for core support.
The content of this publication is solely our responsibility and does not necessarily represent the official views of the NIH.
We thank Carol Ringelberg (Geisel School of Medicine at Dartmouth) for microarray data processing work that was carried out at Dartmouth Medical School in the Genomics Shared Resource. We also thank Fred Ausubel (Harvard Medical School), Daniel Wozniak (The Ohio State University College of Medicine), and Dennis Ohman (Virginia Commonwealth University School of Medicine) for generously providing strains. We acknowledge Gary Heussler (UCSD Division of Biological Sciences) and Jack Hammond for providing plasmids and Jong-In Hong (Seoul National University) for providing the PyDPA probe.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00794-18.
REFERENCES
- 1.Acosta N, Whelan FJ, Somayaji R, Poonja A, Surette MG, Rabin HR, Parkins MD. 2017. The evolving cystic fibrosis microbiome: a comparative cohort study spanning 16 years. Ann ATS 14:1288–1297. doi: 10.1513/AnnalsATS.201609-668OC. [DOI] [PubMed] [Google Scholar]
- 2.Willger SD, Grim SL, Dolben EL, Shipunova A, Hampton TH, Morrison HG, Filkins LM, O‘Toole GA, Moulton LA, Ashare A, Sogin ML, Hogan DA. 2014. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome 2:40. doi: 10.1186/2049-2618-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogan DA, Willger SD, Dolben EL, Hampton TH, Stanton BA, Morrison HG, Sogin ML, Czum J, Ashare A. 2016. Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLoS One 11:e0149998. doi: 10.1371/journal.pone.0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delhaes L, Monchy S, Fréalle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, Sime-Ngando T, Chabé M, Viscogliosi E. 2012. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One 7:e36313. doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkins MD, Floto RA. 2015. Emerging bacterial pathogens and changing concepts of bacterial pathogenesis in cystic fibrosis. J Cyst Fibros 14:293–304. doi: 10.1016/j.jcf.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Bos LD, Meinardi S, Blake D, Whiteson K. 2016. Bacteria in the airways of patients with cystic fibrosis are genetically capable of producing VOCs in breath. J Breath Res 10:047103. doi: 10.1088/1752-7163/10/4/047103. [DOI] [PubMed] [Google Scholar]
- 7.Phan J, Meinardi S, Barletta B, Blake DR, Whiteson K. 2017. Stable isotope profiles reveal active production of VOCs from human-associated microbes. J Breath Res 11:017101. doi: 10.1088/1752-7163/aa5833. [DOI] [PubMed] [Google Scholar]
- 8.Montuschi P, Paris D, Melck D, Lucidi V, Ciabattoni G, Raia V, Calabrese C, Bush A, Barnes PJ, Motta A. 2012. NMR spectroscopy metabolomic profiling of exhaled breath condensate in patients with stable and unstable cystic fibrosis. Thorax 67:222–228. doi: 10.1136/thoraxjnl-2011-200072. [DOI] [PubMed] [Google Scholar]
- 9.Jorgensen F, Bally M, Chapon-Herve V, Michel G, Lazdunski A, Williams P, Stewart GS. 1999. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 145:835–844. doi: 10.1099/13500872-145-4-835. [DOI] [PubMed] [Google Scholar]
- 10.Funabashi H, Ishikawa M, Mie M, Takahashi F, Yanagida Y, Aizawa M, Kobatake E. 2005. Electrochemical evaluation of cellular physiological status under stress in Escherichia coli with the rpoS-lacZ reporter gene. Biotechnol Bioeng 90:509–515. doi: 10.1002/bit.20459. [DOI] [PubMed] [Google Scholar]
- 11.He MX, Wu B, Shui ZX, Hu QC, Wang WG, Tan FR, Tang XY, Zhu QL, Pan K, Li Q, Su XH. 2012. Transcriptome profiling of Zymomonas mobilis under ethanol stress. Biotechnol Biofuels 5:75. doi: 10.1186/1754-6834-5-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen AI, Dolben EF, Okegbe C, Harty CE, Golub Y, Thao S, Ha DG, Willger SD, O'Toole GA, Harwood CS, Dietrich LE, Hogan DA. 2014. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog 10:e1004480. doi: 10.1371/journal.ppat.1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher M. 1983. The effects of methanol, ethanol, propanol and butanol on bacterial attachment to surfaces. J Gen Microbiol 129:633–641. doi: 10.1099/00221287-129-3-633. [DOI] [Google Scholar]
- 14.Morales DK, Grahl N, Okegbe C, Dietrich LE, Jacobs NJ, Hogan DA. 2013. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. mBio 4:e00526-12. doi: 10.1128/mBio.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tashiro Y, Inagaki A, Ono K, Inaba T, Yawata Y, Uchiyama H, Nomura N. 2014. Low concentrations of ethanol stimulate biofilm and pellicle formation in Pseudomonas aeruginosa. Biosci Biotechnol Biochem 78:178–181. doi: 10.1080/09168451.2014.877828. [DOI] [PubMed] [Google Scholar]
- 16.Smith MG, Des Etages SG, Snyder M. 2004. Microbial synergy via an ethanol-triggered pathway. Mol Cell Biol 24:3874–3884. doi: 10.1128/MCB.24.9.3874-3884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camarena L, Bruno V, Euskirchen G, Poggio S, Snyder M. 2010. Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog 6:e1000834. doi: 10.1371/journal.ppat.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nwugo CC, Arivett BA, Zimbler DL, Gaddy JA, Richards AM, Actis LA. 2012. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS One 7:e51936. doi: 10.1371/journal.pone.0051936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterjee I, Somerville GA, Heilmann C, Sahl HG, Maurer HH, Herrmann M. 2006. Very low ethanol concentrations affect the viability and growth recovery in post-stationary-phase Staphylococcus aureus populations. Appl Environ Microbiol 72:2627–2636. doi: 10.1128/AEM.72.4.2627-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiester SE, Actis LA. 2013. Stress responses in the opportunistic pathogen Acinetobacter baumannii. Future Microbiol 8:353–365. doi: 10.2217/fmb.12.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flynn JL, Ohman DE. 1988. Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J Bacteriol 170:1452–1460. doi: 10.1128/jb.170.4.1452-1460.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baynham PJ, Wozniak DJ. 1996. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol Microbiol 22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 23.Martin DW, Schurr MJ, Yu H, Deretic V. 1994. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to sigma E and stress response. J Bacteriol 176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, Deretic V. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol 178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malhotra S, Limoli DH, English AE, Parsek MR, Wozniak DJ. 2018. Mixed communities of mucoid and nonmucoid Pseudomonas aeruginosa exhibit enhanced resistance to host antimicrobials. mBio 9:e00275-18. doi: 10.1128/mBio.00275-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovacikova G, Skorupski K. 2002. The alternative sigma factor sigma E plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect Immun 70:5355–5362. doi: 10.1128/IAI.70.10.5355-5362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haines-Menges B, Whitaker WB, Boyd EF. 2014. Alternative sigma factor RpoE is important for Vibrio parahaemolyticus cell envelope stress response and intestinal colonization. Infect Immun 82:3667–3677. doi: 10.1128/IAI.01854-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rezuchova B, Homerova D, Sevcikova B, Novakova R, Feckova L, Roberts M, Kormanec J. 2013. Phenotypic analysis of Salmonella enterica serovar Typhimurium rpoE mutants encoding RNA polymerase extracytoplasmic stress response sigma factor sigma E with altered promoter specificity. Arch Microbiol 195:27–36. doi: 10.1007/s00203-012-0843-9. [DOI] [PubMed] [Google Scholar]
- 29.Palonen E, Lindstrom M, Somervuo P, Korkeala H. 2013. Alternative sigma factor sigmaE has an important role in stress tolerance of Yersinia pseudotuberculosis IP32953. Appl Environ Microbiol 79:5970–5977. doi: 10.1128/AEM.01891-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishra MN, Kumar S, Gupta N, Kaur S, Gupta A, Tripathi AK. 2011. An extracytoplasmic function sigma factor cotranscribed with its cognate anti-sigma factor confers tolerance to NaCl, ethanol and methylene blue in Azospirillum brasilense Sp7. Microbiology 157:988–999. doi: 10.1099/mic.0.046672-0. [DOI] [PubMed] [Google Scholar]
- 31.Bordes P, Lavatine L, Phok K, Barriot R, Boulanger A, Castanie-Cornet MP, Dejean G, Lauber E, Becker A, Arlat M, Gutierrez C. 2011. Insights into the extracytoplasmic stress response of Xanthomonas campestris pv. campestris: role and regulation of sigma E-dependent activity. J Bacteriol 193:246–264. doi: 10.1128/JB.00884-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boucher JC, Yu H, Mudd MH, Deretic V. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun 65:3838–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. 2008. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigma E in Escherichia coli by both direct and indirect mechanisms. Mol Microbiol 67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- 35.Amar A, Pezzoni M, Pizarro RA, Costa CS. 2018. New envelope stress factors involved in sigma(E) activation and conditional lethality of rpoE mutations in Salmonella enterica. Microbiology 164:1293–1307. doi: 10.1099/mic.0.000701. [DOI] [PubMed] [Google Scholar]
- 36.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266:5980–5990. [PubMed] [Google Scholar]
- 37.Cashel M, Kalbacher B. 1970. The control of ribonucleic acid synthesis in Escherichia coli. V. Characterization of a nucleotide associated with the stringent response. J Biol Chem 245:2309–2318. [PubMed] [Google Scholar]
- 38.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. 2004. Regulation through the secondary channel–structural framework for ppGpp-DksA synergism during transcription. Cell 118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 40.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuster M, Greenberg EP. 2007. Early activation of quorum sensing in Pseudomonas aeruginosa reveals the architecture of a complex regulon. BMC Genomics 8:287. doi: 10.1186/1471-2164-8-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan J, Doing G, Lewis KA, Price CE, Chen KM, Cady KC, Perchuk B, Laub MT, Hogan DA, Greene CS. 2017. Unsupervised extraction of stable expression signatures from public compendia with an ensemble of neural networks. Cell Syst 5:63–71. doi: 10.1016/j.cels.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Djonovic S, Urbach JM, Drenkard E, Bush J, Feinbaum R, Ausubel JL, Traficante D, Risech M, Kocks C, Fischbach MA, Priebe GP, Ausubel FM. 2013. Trehalose biosynthesis promotes Pseudomonas aeruginosa pathogenicity in plants. PLoS Pathog 9:e1003217. doi: 10.1371/journal.ppat.1003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vulic M, Kolter R. 2002. Alcohol-induced delay of viability loss in stationary-phase cultures of Escherichia coli. J Bacteriol 184:2898–2905. doi: 10.1128/JB.184.11.2898-2905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maruta K, Mitsuzumi H, Nakada T, Kubota M, Chaen H, Fukuda S, Sugimoto T, Kurimoto M. 1996. Cloning and sequencing of a cluster of genes encoding novel enzymes of trehalose biosynthesis from thermophilic archaebacterium Sulfolobus acidocaldarius. Biochim Biophys Acta 1291:177–181. doi: 10.1016/S0304-4165(96)00082-7. [DOI] [PubMed] [Google Scholar]
- 46.Freeman BC, Chen C, Beattie GA. 2010. Identification of the trehalose biosynthetic loci of Pseudomonas syringae and their contribution to fitness in the phyllosphere. Environ Microbiol 12:1486–1497. doi: 10.1111/j.1462-2920.2010.02171.x. [DOI] [PubMed] [Google Scholar]
- 47.Wolf A, Kramer R, Morbach S. 2003. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Mol Microbiol 49:1119–1134. doi: 10.1046/j.1365-2958.2003.03625.x. [DOI] [PubMed] [Google Scholar]
- 48.Cardoso FS, Castro RF, Borges N, Santos H. 2007. Biochemical and genetic characterization of the pathways for trehalose metabolism in Propionibacterium freudenreichii, and their role in stress response. Microbiology 153:270–280. doi: 10.1099/mic.0.29262-0. [DOI] [PubMed] [Google Scholar]
- 49.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruhal R, Kataria R, Choudhury B. 2013. Trends in bacterial trehalose metabolism and significant nodes of metabolic pathway in the direction of trehalose accumulation. Microb Biotechnol 6:493–502. doi: 10.1111/1751-7915.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulz S, Eckweiler D, Bielecka A, Nicolai T, Franke R, Dotsch A, Hornischer K, Bruchmann S, Duvel J, Haussler S. 2015. Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog 11:e1004744. doi: 10.1371/journal.ppat.1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tart AH, Wolfgang MC, Wozniak DJ. 2005. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol 187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Damron FH, Qiu D, Yu HD. 2009. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J Bacteriol 191:2285–2295. doi: 10.1128/JB.01490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Damron FH, Owings JP, Okkotsu Y, Varga JJ, Schurr JR, Goldberg JB, Schurr MJ, Yu HD. 2012. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J Bacteriol 194:1317–1330. doi: 10.1128/JB.06105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hengge-Aronis R, Klein W, Lange R, Rimmele M, Boos W. 1991. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J Bacteriol 173:7918–7924. doi: 10.1128/jb.173.24.7918-7924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood LF, Leech AJ, Ohman DE. 2006. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol 62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- 57.Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. 2008. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology 154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wood LF, Ohman DE. 2009. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol 72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 59.Damron FH, Yu HD. 2011. Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J Bacteriol 193:286–291. doi: 10.1128/JB.01132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Damron FH, Goldberg JB. 2012. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol Microbiol 84:595–607. doi: 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schurr MJ, Yu H, Martinez-Salazar JM, Boucher JC, Deretic V. 1996. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol 178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathee K, McPherson CJ, Ohman DE. 1997. Posttranslational control of the algT (algU)-encoded sigma 22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol 179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cezairliyan BO, Sauer RT. 2009. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol 72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A 90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rowen DW, Deretic V. 2000. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol Microbiol 36:314–327. doi: 10.1046/j.1365-2958.2000.01830.x. [DOI] [PubMed] [Google Scholar]
- 66.Ohman DE, Chakrabarty AM. 1981. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun 33:142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DeVries CA, Ohman DE. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol 176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chand NS, Clatworthy AE, Hung DT. 2012. The two-component sensor KinB acts as a phosphatase to regulate Pseudomonas aeruginosa virulence. J Bacteriol 194:6537–6547. doi: 10.1128/JB.01168-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci U S A 102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mallik P, Paul BJ, Rutherford ST, Gourse RL, Osuna R. 2006. DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J Bacteriol 188:5775–5782. doi: 10.1128/JB.00276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu J, Xie J. 2009. Magic spot: (p) ppGpp. J Cell Physiol 220:297–302. doi: 10.1002/jcp.21797. [DOI] [PubMed] [Google Scholar]
- 73.Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T. 2011. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol 80:1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gentry DR, Cashel M. 1996. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol 19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- 75.Montero M, Rahimpour M, Viale AM, Almagro G, Eydallin G, Sevilla Á, Cánovas M, Bernal C, Lozano AB, Muñoz FJ, Baroja-Fernández E, Bahaji A, Mori H, Codoñer FM, Pozueta-Romero J. 2014. Systematic production of inactivating and non-inactivating suppressor mutations at the relA locus that compensate the detrimental effects of complete spoT loss and affect glycogen content in Escherichia coli. PLoS One 9:e106938. doi: 10.1371/journal.pone.0106938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, de Crécy-Lagard V. 2011. Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol Microbiol 79:700–715. doi: 10.1111/j.1365-2958.2010.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jude F, Kohler T, Branny P, Perron K, Mayer MP, Comte R, van Delden C. 2003. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J Bacteriol 185:3558–3566. doi: 10.1128/JB.185.12.3558-3566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.D'Souza-Ault MR, Smith LT, Smith GM. 1993. Roles of N-acetylglutaminylglutamine amide and glycine betaine in adaptation of Pseudomonas aeruginosa to osmotic stress. Appl Environ Microbiol 59:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Kievit TR, Kakai Y, Register JK, Pesci EC, Iglewski BH. 2002. Role of the Pseudomonas aeruginosa Las and Rhl quorum-sensing systems in RhlI regulation. FEMS Microbiol Lett 212:101–106. doi: 10.1111/j.1574-6968.2002.tb11251.x. [DOI] [PubMed] [Google Scholar]
- 80.Tan J, Huyck M, Hu D, Zelaya RA, Hogan DA, Greene CS. 2017. ADAGE signature analysis: differential expression analysis with data-defined gene sets. BMC Bioinformatics 18:512. doi: 10.1186/s12859-017-1905-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tan J, Hammond JH, Hogan DA, Greene CS. 2016. ADAGE-based integration of publicly available Pseudomonas aeruginosa gene expression data with denoising autoencoders illuminates microbe-host interactions. mSystems 1:e00025-15. doi: 10.1128/mSystems.00025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Costanzo A, Ades SE. 2006. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigma E, by guanosine 3',5'-bispyrophosphate (ppGpp). J Bacteriol 188:4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cugini C, Morales DK, Hogan DA. 2010. Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 156:3096–3107. doi: 10.1099/mic.0.037911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schafhauser J, Lepine F, McKay G, Ahlgren HG, Khakimova M, Nguyen D. 2014. The stringent response modulates 4-hydroxy-2-alkylquinoline biosynthesis and quorum-sensing hierarchy in Pseudomonas aeruginosa. J Bacteriol 196:1641–1650. doi: 10.1128/JB.01086-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu X, Yu H, Zhang D, Xiong J, Qiu J, Xin R, He X, Sheng H, Cai W, Jiang L, Zhang K, Hu X. 2016. Role of ppGpp in Pseudomonas aeruginosa acute pulmonary infection and virulence regulation. Microbiol Res 192:84–95. doi: 10.1016/j.micres.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 86.Erickson DL, Lines JL, Pesci EC, Venturi V, Storey DG. 2004. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect Immun 72:5638–5645. doi: 10.1128/IAI.72.10.5638-5645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baysse C, Cullinane M, Denervaud V, Burrowes E, Dow JM, Morrissey JP, Tam L, Trevors JT, O'Gara F. 2005. Modulation of quorum sensing in Pseudomonas aeruginosa through alteration of membrane properties. Microbiology 151:2529–2542. doi: 10.1099/mic.0.28185-0. [DOI] [PubMed] [Google Scholar]
- 88.Branny P, Pearson JP, Pesci EC, Kohler T, Iglewski BH, Van Delden C. 2001. Inhibition of quorum sensing by a Pseudomonas aeruginosa DksA homologue. J Bacteriol 183:1531–1539. doi: 10.1128/JB.183.5.1531-1539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schnider-Keel U, Lejbolle KB, Baehler E, Haas D, Keel C. 2001. The sigma factor AlgU (AlgT) controls exopolysaccharide production and tolerance towards desiccation and osmotic stress in the biocontrol agent Pseudomonas fluorescens CHA0. Appl Environ Microbiol 67:5683–5693. doi: 10.1128/AEM.67.12.5683-5693.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Freeman BC, Chen C, Yu X, Nielsen L, Peterson K, Beattie GA. 2013. Physiological and transcriptional responses to osmotic stress of two Pseudomonas syringae strains that differ in epiphytic fitness and osmotolerance. J Bacteriol 195:4742–4752. doi: 10.1128/JB.00787-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu X, Lund SP, Greenwald JW, Records AH, Scott RA, Nettleton D, Lindow SE, Gross DC, Beattie GA. 2014. Transcriptional analysis of the global regulatory networks active in Pseudomonas syringae during leaf colonization. mBio 5:e01683-14. doi: 10.1128/mBio.01683-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Markel E, Stodghill P, Bao Z, Myers CR, Swingle B. 2016. AlgU controls expression of virulence genes in Pseudomonas syringae pv. tomato DC3000. J Bacteriol 198:2330–2344. doi: 10.1128/JB.00276-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elbein AD, Pan YT, Pastuszak I, Carroll D. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 94.De Virgilio C, Hottiger T, Dominguez J, Boller T, Wiemken A. 1994. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. I. Genetic evidence that trehalose is a thermoprotectant. Eur J Biochem 219:179–186. doi: 10.1111/j.1432-1033.1994.tb19928.x. [DOI] [PubMed] [Google Scholar]
- 95.Hottiger T, De Virgilio C, Hall MN, Boller T, Wiemken A. 1994. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur J Biochem 219:187–193. doi: 10.1111/j.1432-1033.1994.tb19929.x. [DOI] [PubMed] [Google Scholar]
- 96.Kandror O, DeLeon A, Goldberg AL. 2002. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc Natl Acad Sci U S A 99:9727–9732. doi: 10.1073/pnas.142314099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singer MA, Lindquist S. 1998. Thermotolerance in Saccharomyces cerevisiae: the yin and yang of trehalose. Trends Biotechnol 16:460–468. doi: 10.1016/S0167-7799(98)01251-7. [DOI] [PubMed] [Google Scholar]
- 98.Thammahong A, Puttikamonkul S, Perfect JR, Brennan RG, Cramer RA. 2017. Central role of the trehalose biosynthesis pathway in the pathogenesis of human fungal infections: opportunities and challenges for therapeutic development. Microbiol Mol Biol Rev 81:e00053-16. doi: 10.1128/MMBR.00053-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bosch S, de Beaurepaire L, Allard M, Mosser M, Heichette C, Chretien D, Jegou D, Bach JM. 2016. Trehalose prevents aggregation of exosomes and cryodamage. Sci Rep 6:36162. doi: 10.1038/srep36162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kurz M, Burch AY, Seip B, Lindow SE, Gross H. 2010. Genome-driven investigation of compatible solute biosynthesis pathways of Pseudomonas syringae pv. syringae and their contribution to water stress tolerance. Appl Environ Microbiol 76:5452–5462. doi: 10.1128/AEM.00686-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Waite RD, Papakonstantinopoulou A, Littler E, Curtis MA. 2005. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J Bacteriol 187:6571–6576. doi: 10.1128/JB.187.18.6571-6576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Waite RD, Paccanaro A, Papakonstantinopoulou A, Hurst JM, Saqi M, Littler E, Curtis MA. 2006. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7:162. doi: 10.1186/1471-2164-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chandra G, Chater KF, Bornemann S. 2011. Unexpected and widespread connections between bacterial glycogen and trehalose metabolism. Microbiology 157:1565–1572. doi: 10.1099/mic.0.044263-0. [DOI] [PubMed] [Google Scholar]
- 104.Park SC, Pham BP, Van Duyet L, Jia B, Lee S, Yu R, Han SW, Yang JK, Hahm KS, Cheong GW. 2008. Structural and functional characterization of osmotically inducible protein C (OsmC) from Thermococcus kodakaraensis KOD1. Biochim Biophys Acta 1784:783–788. doi: 10.1016/j.bbapap.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 105.Pelzer A, Polen T, Funken H, Rosenau F, Wilhelm S, Bott M, Jaeger KE. 2014. Subtilase SprP exerts pleiotropic effects in Pseudomonas aeruginosa. Microbiologyopen 3:89–103. doi: 10.1002/mbo3.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rodriguez-Rojas A, Blazquez J. 2009. The Pseudomonas aeruginosa pfpI gene plays an antimutator role and provides general stress protection. J Bacteriol 191:844–850. doi: 10.1128/JB.01081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mitchell JJ, Lucas-Lenard JM. 1980. The effect of alcohols on guanosine 5'-diphosphate-3'-diphosphate metabolism in stringent and relaxed Escherichia coli. J Biol Chem 255:6307–6313. [PubMed] [Google Scholar]
- 108.Haft RJ, Keating DH, Schwaegler T, Schwalbach MS, Vinokur J, Tremaine M, Peters JM, Kotlajich MV, Pohlmann EL, Ong IM, Grass JA, Kiley PJ, Landick R. 2014. Correcting direct effects of ethanol on translation and transcription machinery confers ethanol tolerance in bacteria. Proc Natl Acad Sci U S A 111:E2576–E2585. doi: 10.1073/pnas.1401853111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for Enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 113.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Friedman L, Kolter R. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol 186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jackson AA, Gross MJ, Daniels EF, Hampton TH, Hammond JH, Vallet-Gely I, Dove SL, Stanton BA, Hogan DA. 2013. Anr and its activation by PlcH activity in Pseudomonas aeruginosa host colonization and virulence. J Bacteriol 195:3093–3104. doi: 10.1128/JB.02169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Irizarry RA, Hobbs B, Collin F, Beazer BY, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 117.Butts C. 2008. network: a package for managing relational data in R. J Statistical Software 24:1–36. doi: 10.18637/jss.v024.i02. [DOI] [Google Scholar]
- 118.Butts C. 2015. network: classes for relational data, vR package version 1.13.0.1. https://CRAN.R-project.org/package=network.
- 119.Barret Schloerke JC, Cook D, Briatte F, Marbach M, Thoen E, Elberg A, Larmarange J. 2018. GGally: extension to 'ggplot2', v1.4.0. https://ggobi.github.io/ggally.
- 120.Wickham H. 2016. ggplot2: elegant graphics for data analysis, 1st ed Springer-Verlag, New York, NY. [Google Scholar]
- 121.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.d'Enfert C, Fontaine T. 1997. Molecular characterization of the Aspergillus nidulans treA gene encoding an acid trehalase required for growth on trehalose. Mol Microbiol 24:203–216. doi: 10.1046/j.1365-2958.1997.3131693.x. [DOI] [PubMed] [Google Scholar]
- 123.Puttikamonkul S, Willger SD, Grahl N, Perfect JR, Movahed N, Bothner B, Park S, Paderu P, Perlin DS, Cramer RA Jr. 2010. Trehalose 6-phosphate phosphatase is required for cell wall integrity and fungal virulence but not trehalose biosynthesis in the human fungal pathogen Aspergillus fumigatus. Mol Microbiol 77:891–911. doi: 10.1111/j.1365-2958.2010.07254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lagosky PA, Chang FN. 1978. The extraction of guanosine 5'-diphosphate, 3'-diphosphate (ppGpp) from Escherichia coli using low pH reagents: a reevaluation. Biochem Biophys Res Commun 84:1016–1024. doi: 10.1016/0006-291X(78)91685-6. [DOI] [PubMed] [Google Scholar]
- 125.Rhee HW, Lee CR, Cho SH, Song MR, Cashel M, Choy HE, Seok YJ, Hong JI. 2008. Selective fluorescent chemosensor for the bacterial alarmone (p)ppGpp. J Am Chem Soc 130:784–785. doi: 10.1021/ja0759139. [DOI] [PubMed] [Google Scholar]
- 126.Malke H. 1993. Jeffrey H. Miller, A short course in bacterial genetics—a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor 1992. Cold Spring Harbor Laboratory Press. J Basic Microbiol 33:1. doi: 10.1002/jobm.3620330412. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.