The two lantibiotics nisin and subtilin are produced by Lactococcus lactis and Bacillus subtilis, respectively. Both peptides have strong antimicrobial activity against Gram-positive bacteria, and therefore, appropriate protection mechanisms are required for the producing strains. To prevent toxicity of their own lantibiotic, both bacteria express immunity proteins, called SpaI and NisI, and in addition, ABC transporters SpaFEG and NisFEG. Whereas it has been shown that the ABC transporters protect the producing strains by transporting the toxic peptides to the extracellular space, the exact mode of action and the physiological function of the lipoproteins during immunity are still unknown. Understanding the exact role of lantibiotic immunity proteins is of major importance for improving production rates and for the design of newly engineered peptide antibiotics. Here, we show (i) the specificity of each lipoprotein for its own lantibiotic, (ii) the specific physical interaction of subtilin with its lipoprotein SpaI, (iii) the physiological function of SpaI in protecting the cellular membrane, and (iv) the importance of the C-terminal part of subtilin for its interaction with SpaI.
KEYWORDS: antibiotic resistance, Bacillus subtilis, entianin, immunity, LILBID, Lactococcus lactis, lantibiotics, nisin, subtilin
ABSTRACT
Lantibiotics subtilin and nisin are produced by Bacillus subtilis and Lactococcus lactis, respectively. To prevent toxicity of their own lantibiotic, both bacteria express specific immunity proteins, called SpaI and NisI. In addition, ABC transporters SpaFEG and NisFEG prevent lantibiotic toxicity by transporting the respective peptides to the extracellular space. Although the three-dimensional structures of SpaI and NisI have been solved, very little is known about the molecular function of either lipoprotein. Using laser-induced liquid bead ion desorption (LILBID)–mass spectrometry, we show here that subtilin interacts with SpaI monomers. The expression of either SpaI or NisI in a subtilin-nonproducing B. subtilis strain resulted in the respective strain being more resistant against either subtilin or nisin. Furthermore, pore formation provided by subtilin and nisin was prevented specifically upon the expression of either SpaI or NisI. As shown with a nisin-subtilin hybrid molecule, the C-terminal part of subtilin but not any particular lanthionine ring was needed for SpaI-mediated immunity. With respect to growth, SpaI provided less immunity against subtilin than is provided by the ABC transporter SpaFEG. However, SpaI prevented pore formation much more efficiently than SpaFEG. Taken together, our data show the physiological function of SpaI as a fast immune response to protect the cellular membrane.
IMPORTANCE The two lantibiotics nisin and subtilin are produced by Lactococcus lactis and Bacillus subtilis, respectively. Both peptides have strong antimicrobial activity against Gram-positive bacteria, and therefore, appropriate protection mechanisms are required for the producing strains. To prevent toxicity of their own lantibiotic, both bacteria express immunity proteins, called SpaI and NisI, and in addition, ABC transporters SpaFEG and NisFEG. Whereas it has been shown that the ABC transporters protect the producing strains by transporting the toxic peptides to the extracellular space, the exact mode of action and the physiological function of the lipoproteins during immunity are still unknown. Understanding the exact role of lantibiotic immunity proteins is of major importance for improving production rates and for the design of newly engineered peptide antibiotics. Here, we show (i) the specificity of each lipoprotein for its own lantibiotic, (ii) the specific physical interaction of subtilin with its lipoprotein SpaI, (iii) the physiological function of SpaI in protecting the cellular membrane, and (iv) the importance of the C-terminal part of subtilin for its interaction with SpaI.
INTRODUCTION
The investigation and further development of new antimicrobially active substances has become a challenging field due to the rising amount of multidrug resistances in human pathogens like methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE). Furthermore, negative effects of antibiotics on the bacterial flora due to long-term application cannot be ruled out at this point (1, 2). Besides the commonly used broad-spectrum antibiotics, ribosomally synthesized and posttranslationally modified peptides (RiPPs) are being considered as an alternative (3). In contrast to nonribosomal antibiotics, RiPPs are accessible to easy genetic manipulation to improve their antimicrobial activities.
Ribosomally synthesized peptides include the large group of lantibiotics, which are produced by Gram-positive bacteria. Lantibiotics have been intensively investigated during the last decades. The characteristic nonproteinogenic amino acid 3,3′-thiodialanine, also called lanthionine, represents the monosulfide analog of cysteine and is composed of two alanine residues which in turn are cross-linked on their β-carbon atoms via a thioether. The chemical formula of lanthionine is [HOOC-CH(NH2)-CH2-S-CH2-CH(NH2)-COOH]. Due to their composition, the lanthionine-containing peptides (lantibiotics) exhibit high stability and also unique biological activities. Based on their characteristic chemical structure and their antibiotic activity, the nomenclature proposed by Schnell et al. (4) summarized these peptides as lantibiotics (lanthionine-containing antibiotics). Epidermin was the first lantibiotic for which the ribosomal synthesis was proven (4). Shortly after, there was a great increase in the published number of structural genes encoding other lantibiotics, such as spaS for subtilin (5), nisA for nisin (6, 7), gdmA for gallidermin (8, 9), pepA for Pep 5 (10), and cinA for cinnamycin (11). Today, more than 95 different lantibiotics have been identified, and many of them are potent candidates for antibiotic applications (12, 13).
Nisin represents the most prominent member of the lantibiotics, and this peptide is frequently employed as a food preservative, better known as E234 (14, 15). Major advantages of lantibiotics are their stability against heat and oxidation and, for their application in food industry, their early proteolytic degradation within the human stomach. However, since lantibiotics are unable to pass the human alimentary tract, their medical application is restricted compared to conventional antibiotics.
The biosynthesis of the linear lantibiotics nisin and subtilin occurs at multimeric protein complexes (lanthionine synthetase). These localize at the cellular membrane and consist of SpaB (NisB), SpaC (NisC), and the ABC transporter SpaT (NisT). All three proteins are embedded as dimers in the respective lanthionine synthetase complexes (16–18). As shown for nisin, NisB encodes the lanthionine dehydratase that eliminates water from either serine or threonine (19), whereas the lanthionine cyclase NisC catalyzes the biosynthesis of a sulfur bridge (20, 21). As already postulated by Schnell et al. (4), the leader peptide was shown to be essential for lantibiotic maturation (22). Whereas the nisin leader peptide is removed by the specific protease NisP (23, 24), several proteases are needed for the removal of the leader peptide during subtilin biosynthesis (25).
Generally, the respective Gram-positive bacteria produce lantibiotics in a nanomolar range to provide growth advantages against bacterial competitors in close proximity. However, as lantibiotics would also be active against their producing organisms, these strains possess immunity genes that protect the lantibiotic producer from the toxic effects of its own antibiotic. These immunity genes are specific for lantibiotics and, thus, are different from various broad-range protection systems, such as the Psd, Bce, or LiaI systems that are expressed by many bacteria for their defense against toxic peptides (26–28).
As first shown for nisin and subtilin, two kinds of immunity proteins were discovered, comprising a lipoprotein (SpaI for subtilin-like lantibiotics and NisI for nisin) and an ABC transporter (SpaFEG and NisFEG) (29, 30). This is different for epidermin- and gallidermin-producing strains (Staphylococcus epidermidis and Staphylococcusgallinarum), which contain unique genes called epiH and gdmH (LanH). These genes encode accessory factors for the ATP-binding cassette transporters, and gdmH provides strong immunity against gallidermin in combined expression with gdmT (31). LanH contains three transmembrane helices, and a similar protein has also been described for the lantibiotic gene cluster of nukacin ISK-1 (32, 33). For Pep5 immunity, the immunity protein PepI seems to be completely different from the lantibiotic immunity genes described above. PepI is located outside the cytoplasm and consists of 69 amino acids, of which the 20 C-terminal amino acids containing 8 positively charged residues are needed for immunity. The molecular mechanism of PepI immunity has not been solved so far, but due to the fact that PepI and lantibiotic Pep5 contain similar amounts of positively charged amino acids, a mechanism is discussed where PepI competes with Pep5 for a so-far-unknown anionic compound which is needed for Pep5-mediated pore formation (34).
In the case of nisin produced by Lactococcus lactis and subtilin produced by Bacillus subtilis, full self-protection is only achieved when NisI and the NisFEG transporter are both expressed, although each system is able to confer partial immunity (29, 35–40). The question of whether NisI (SpaI)- and NisFEG (SpaFEG)-mediated immunities are additive or cooperative still remains controversial (36, 37, 41). The LanFEG-type ABC transporters generally consist of two permeases, LanE and LanG, which both contain six transmembrane helices, as well as the ATPase LanF (42). Immunity mediated by the ABC transporter is based on the transport of the lantibiotic molecules to the extracellular space (37), thereby causing a change in the distribution equilibrium between the target-associated and the free peptide (43, 37, 38, 44, 45). The exact immunity mechanism of LanI is still poorly understood. Nevertheless, it is known that LanI-type immunity proteins are proceeded by an N-terminal leader sequence and a consensus lipobox motif, L[A/S][G/A]C (46), in front of the core peptide (47, 24, 29, 48).
Generally, four different mechanisms for SpaI-mediated immunity are conceivable: (i) protection of the membrane from lantibiotic insertion by binding and sequestering the peptides, (ii) protection of the membrane from lantibiotic insertion by binding and sequestering of lantibiotic-lipid II complexes prior to pore formation, (iii) a shielding mechanism that closes subtilin/lipid II pores, and (iv) a mechanism where the lipoprotein could also act as a substrate-binding protein for the ABC transporter LanFEG (49). All these mechanisms presume a direct interaction of LanI with the respective lantibiotic, which, however, has only been shown for NisI and nisin so far (50).
The structure of the B. subtilis LanI protein SpaI was recently solved, revealing a novel three-dimensional (3-D) fold (49). Since LanI proteins share very weak sequence homology (identity of ∼11% and similarity of approximately ∼20%) (Fig. S2 in the supplemental material) and, furthermore, differ significantly in their molecular weights (16.8 kDa for SpaI and 25.8 kDa for NisI), it was most surprising that the overall structures of SpaI and NisI were very similar (50). As a unique feature, the lipoprotein NisI is composed of two separate domains with different pI values of 6 (C-terminal domain) and 9 (N-terminal domain). The domains are separated by a linker region. Nisin binds to the C-terminal part of NisI, which is most likely oriented away from the membrane due to its negative charge. In contrast, the more basic N-terminal domain of NisI binds to membranes but does not bind nisin (50).
In this work, we show that the lipoprotein SpaI recognizes the C-terminal part of the antimicrobial peptide and mediates specific immunity against it. Furthermore, we provide evidence that the LanI protein, in contrast to the ABC transporter, provides a fast immunity response within the first seconds to minutes, whereas the protective effect of the ABC transporter occurs with some delay.
RESULTS
Subtilin specifically interacts with the B. subtilis immunity protein SpaI.
Whereas an interaction of nisin from L. lactis with its respective immunity protein NisI was proven previously (39, 50), all attempts to show a physical interaction of the immunity protein SpaI with subtilin were not successful so far, as the different pI values of SpaI and subtilin made it impossible to keep both proteins in solution within the same buffer (49). Currently, only cross-linking experiments suggested a direct in vitro interaction between SpaI and subtilin (38). As an alternative physical method to analyze the interaction of subtilin with its immunity protein SpaI, we applied the recently established biophysical method called laser-induced liquid bead ion desorption (LILBID). During LILBID analysis, biomolecules are desorbed by laser pulses from liquid microdroplets, and by varying the laser intensity, both the masses of entire protein complexes (low laser intensity) and the subunits from the disassembled complex (high laser intensity) can be detected.
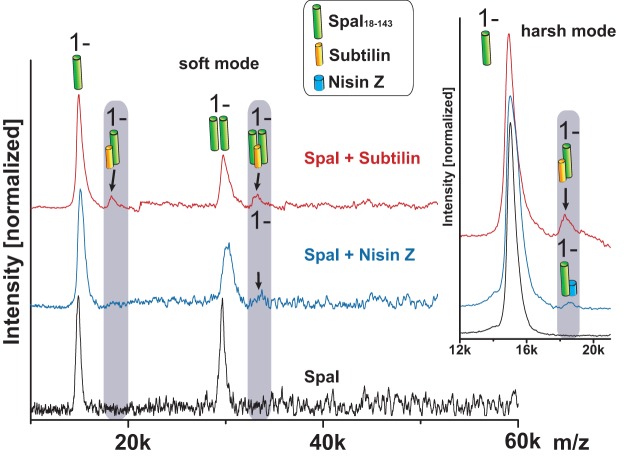
For LILBID experiments, we used the N-terminally truncated SpaI18–143 variant for which the 3-D structure was determined via nuclear magnetic resonance (NMR), as these studies showed that the N-terminal amino acids of SpaI are flexible and, thus, a target for proteolytic degradation (49). SpaI18–143 was measured in the absence of lantibiotic (Fig. 1, black) and in the presence of nisin (Fig. 1, blue) or subtilin (Fig. 1, red). Indeed, the truncated SpaI18–143 protein provided two distinct LILBID signals with comparable intensities, corresponding to the SpaI18–143 monomer of ∼15 kDa and the SpaI18–143 dimer of ∼30 kDa. If subtilin was added, additional signals occurred at ∼18 kDa and ∼33 kDa. The ∼18-kDa signal corresponded to a SpaI-subtilin complex consisting of the SpaI monomer and subtilin. Interestingly, the ∼33-kDa signal corresponded to a complex consisting of the SpaI dimer with only one subtilin molecule. By increasing the laser intensity (harsh mode), the SpaI dimer-subtilin complex could be dissociated, whereas the SpaI monomer-subtilin complex remained stable. The addition of nisin to SpaI18–143 resulted in a potentially weak signal at ∼34 kDa (Fig. 1, SpaI + nisin Z, black arrow), close to the noise signals. To show the significance of the 34-kDa peak, repeated measurements with optimized settings for the detection of higher masses (15 mJ) were performed (Fig. S5). Spectra averaged from 1,000 single measurements confirmed the weak binding of nisin to SpaI. Analysis of SpaI18–143 using LILBID revealed masses/signals indicative of a SpaI18–143 molecule with two (*a) or even three (*b) nisin Z molecules, possibly as a result of a nonspecific/weaker bond. Higher laser intensity (20 mJ) removed double and triple bound nisin-SpaI complexes, and only a single nisin molecule bound to the SpaI monomer remained stable (Fig. S5). These data showed that a significant SpaI-nisin complex was built, although its amount was much less than that of SpaI-subtilin.
FIG 1.
LILBID analysis of the interaction between the truncated immunity protein SpaI18–143 and class I lantibiotics subtilin and nisin Z. Compendium of the measurements of SpaI18–143 (black), SpaI18–143 plus nisin Z (blue), and SpaI18–143 plus subtilin (red). The zoomed area points out the complex of SpaI18–143 plus subtilin (red) or nisin Z (blue). The spectra shown were recorded at low laser intensity (15 mJ, soft mode) and high laser intensity (20 mJ, harsh mode). Instrumental settings and protein concentrations were identical for all measurements. Lantibiotics were applied in 3-fold excess (48 µM) over the concentration of SpaI (16 µM), and spectra are averaged out of 500 single measurements.
In summary, our data show the direct interaction of immunity protein SpaI with its ligand subtilin using a physical method, which mimics the native situation more closely than other methods used so far. However, the amount of SpaI-subtilin complexes detected was still low. Due to the method applied, this does not provide any hint to the affinity of subtilin for SpaI, as laser intensities also interfere with the stability of the SpaI-subtilin complex. In conclusion, LILBID analyses clearly confirmed the results of our previous cross-linking analysis (38), showing that the monomeric state of the SpaI protein is sufficient for subtilin binding and no dimeric state is required.
Cellular protection mediated by SpaI and NisI.
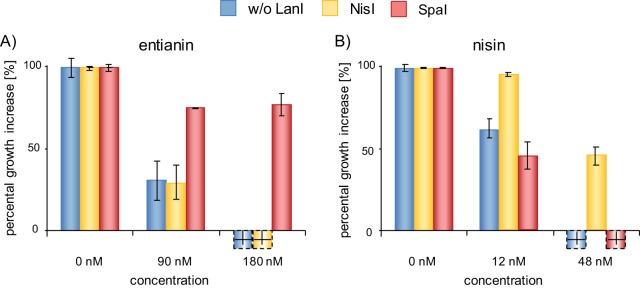
The cross-linking and the LILBID measurements both represent in vitro test systems, and surprisingly, LILBID measurements also showed a very weak interaction between SpaI and nisin. For this reason, we established in vivo test systems to assess the efficiencies of SpaI- and NisI-mediated immunity. The respective immunity proteins were expressed in a non-lantibiotic producer (B. subtilis strain 168) under the regulation of a xylose-inducible promoter. The results in Fig. 2 show the levels of immunity mediated by SpaI (B168.SB2) and NisI (B168.SB1) compared to that of a negative control without LanI (B168.KO). Due to its simplified purification, we applied the recently described subtilin-like lantibiotic entianin, which differs from subtilin in three amino acid positions (6, 15, and 24) (Fig. S1). The EtnI protein of entianin producer B. subtilis DSM 15029 shares sequence similarities of 95% with SpaI. Therefore, it was not surprising that SpaI also mediated specific immunity against entianin (51, 49).
FIG 2.
NisI- and SpaI-mediated immunity against class I lantibiotics entianin and nisin. Percental growth increase after 0.5 h of incubation with entianin (A) and nisin (B) normalized to the results for a control without (w/o) lantibiotic (blue, control strain B168.KO; yellow, NisI-expressing strain B168.SB1; red, SpaI-expressing strain B168.SB2). The hatched bars indicate decreases in optical density, presumably due to cell lysis. Values provided are the means of three independent experiments.
As expected, the negative-control strain B168.KO without LanI showed strong susceptibility against both lantibiotics. The addition of 90 nM entianin and 12 nM nisin already resulted in a drastic effect on growth. In contrast, SpaI (Fig. 2, B168.SB2, red bars) and NisI (Fig. 2, B168.SB1, yellow bars) conferred immunity against the corresponding lantibiotics. The immunity provided by SpaI was directed specifically against subtilin, and that of NisI was particular against nisin. Due to the increased activity of nisin against Bacillus, lower nisin concentrations provided inhibition effects similar to those of subtilin (Fig. 2 and Table S1).
SpaI and NisI prevent pore formation in a specific manner.
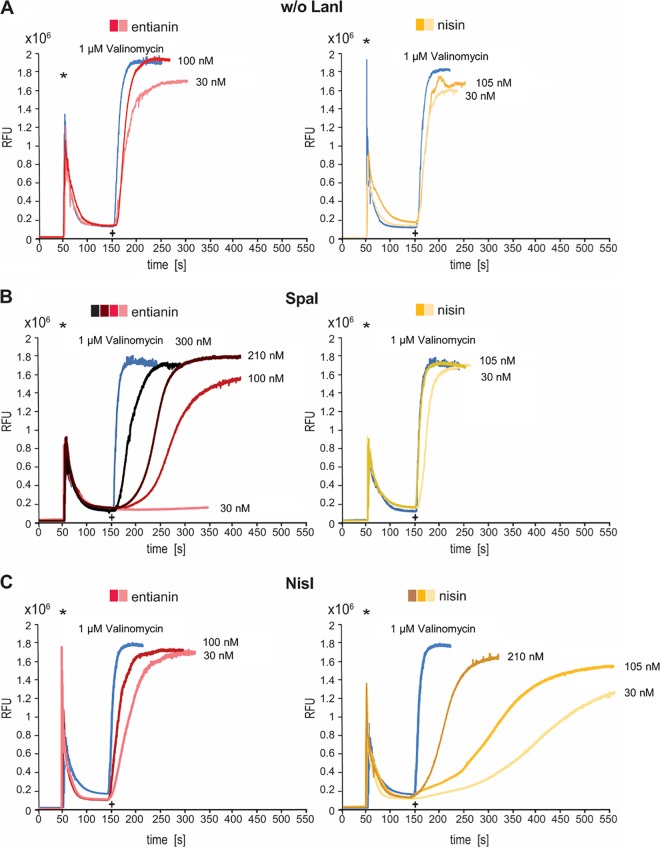
After binding to the cell wall precursor lipid II, the class I lantibiotics nisin and subtilin form stable pores in the membrane of Gram-positive bacteria (52–56). The respective pore complexes consist of eight lantibiotic molecules in combination with four lipid II molecules (57, 58). With a diameter of approximately 2 to 2.5 nm (59), these pore complexes cause the leakage of small molecules and, finally, the collapse of the membrane potential. The resulting breakdown of the membrane potential (60) can be visualized by following the leakage of the fluorescent dye DiSC3(5) (3,3′-dipropylthiadicarbocyanine iodide). Upon its addition, DiSC3(5) inserts into the membrane, accelerated by the membrane potential gradient, thereby quenching its own fluorescence. After the addition of pore-forming peptides, the membrane potential is disturbed and the fluorescent dye is released (61). For more detailed information, see reference 62. As a positive control, membrane dissipation can be followed using the depsipeptide valinomycin, which selectively transports K+ ions across the membrane, thereby destroying the membrane potential and resulting in the release of DiSC3(5).
The addition of entianin, as well as nisin, to B. subtilis cells expressing no lantibiotic immunity genes (B168.KO) led to an immediate breakdown of the membrane potential (Fig. 3A). After the addition of 100 nM entianin, an instant increase in fluorescence, comparable to that caused by valinomycin, was observed. Even entianin concentrations of 30 nM dissipated the membrane to a similar extent. Comparable results were obtained with nisin using concentrations of 105 nM and 30 nM. The expression of SpaI (B168.SB2) provided an increased resistance against entianin. After the addition of 30 nM entianin, almost no increase in fluorescence was observed, and the addition of 100 nM entianin had only a moderate effect. The addition of 210 nM entianin caused maximal relative fluorescence units (RFU) of approximately 1.8 × 106 RFU, comparable to the value obtained using 1 μM valinomycin; however, the slope was significantly lower. Only a concentration of 300 nM entianin led to the immediate release of the fluorescent dye. No increased resistance against nisin was provided by immunity protein SpaI (Fig. 3B). In an experiment performed vice versa, the NisI-expressing strain B168.SB1 tolerated higher nisin concentrations, and even the addition of 210 nM nisin decelerated the fluorescence increase compared to the results for the valinomycin control. In summary, these findings mainly confirmed the growth test results (Fig. 2). Moreover, the DiSC3(5) diffusion assay clearly proved that SpaI and NisI prevent pore formation and, thus, diminish the impact of entianin and nisin on the membrane.
FIG 3.
DiSC3(5) diffusion assay for analysis of SpaI- and NisI-mediated immunity against entianin and nisin. Cells were incubated with the fluorescent dye DiSC3(5), and fluorescence was monitored (emission, 670 nm; excitation, 544 nm). Asterisks indicate the time of 2.5 μM DiSC3(5) addition, and plus signs indicate the time when either entianin (red), nisin (yellow), or valinomycin (blue) was added to control strain B168.KO (A), SpaI-expressing strain B168.SB2 (B), and NisI-expressing strain B168.SB1 (C).
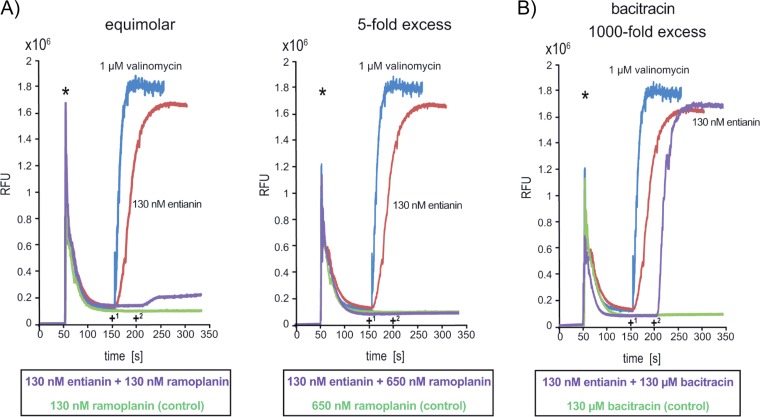
To analyze the importance of lipid II for subtilin- and nisin-mediated pore formation, the lipodepsipeptide ramoplanin (63–66) was applied together with the respective lantibiotics in the DiSC3(5) diffusion assays (Fig. 4). Ramoplanin recognizes the N-acetylmuramic acid (MurNAc)-Ala-Glu pyrophosphate unit of lipid II (67) and, thus, competes with entianin and nisin for the same lipid II binding site (55, 68). Ramoplanin itself does not lead to a breakdown of the membrane potential. When it was applied in equimolar concentrations with entianin (Fig. 4A), the collapse of the membrane potential was prevented to a major extent. After the addition of ramoplanin in a 5-fold excess, the entianin-mediated effect was completely suppressed (Fig. 4A). This clearly indicated the important role of lipid II for the antibiotic activity of subtilin-like lantibiotics.
FIG 4.
Impact of ramoplanin (A) and bacitracin (B) on entianin-mediated breakdown of the membrane potential using the DiSC3(5) diffusion assay. B168.KO (without immunity proteins) cells were incubated with the fluorescent dye DiSC3(5), and fluorescence was monitored (emission, 670 nm; excitation, 544 nm). Asterisks indicate the time of 2.5 μM DiSC3(5) addition. (A) Ramoplanin applied in equimolar concentration with entianin (130 nM, left) and in 5-fold excess (650 nM, right). +1, time when entianin (red), valinomycin (blue), or ramoplanin (green) was added; +2, time of addition of entianin to cells preincubated with ramoplanin (purple). (B) Bacitracin applied in 1,000-fold molar excess with entianin (130 nM). +1, time when entianin (red), valinomycin (blue), or bacitracin (green) was added; +2, time of further addition of entianin to cells preincubated with bacitracin (purple).
To ensure that the suppression of entianin toxicity by ramoplanin was due to its binding to lipid II and not due to any interference with lipid II biosynthesis, we used bacitracin, a cyclic peptide that prevents the recycling of lipid II upon binding to undecaprenyl pyrophosphate (69, 70). The DiSC3(5) diffusion assay clearly showed that bacitracin had no influence on the entianin-mediated breakdown of the membrane potential even if applied in 1,000-fold excess (Fig. 4B). To exclude any competition for the target site, entianin was added 50 s after the addition of bacitracin.
SpaI recognizes the C-terminal part of the lantibiotic.
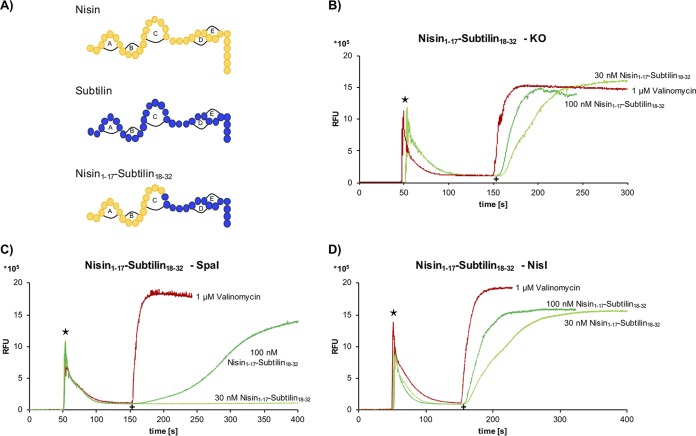
To identify structural constraints that are important for the specific recognition by the respective immunity proteins, we used the previously described subtilin-nisin hybrid peptide consisting of the N-terminal part of nisin up to amino acid position 17 and the C-terminal part of subtilin comprising amino acid positions 18 to 32 (nisin1–17-subtilin18–32) (Fig. 5A) (71).
FIG 5.
Influence of nisin1–17-subtilin18–32 peptide on SpaI-/NisI-mediated immunity in a DiSC3(5) diffusion assay. (A) Primary structures of the lantibiotics tested in this study. (B) Control strain B168.KO (without immunity proteins) was incubated with DiSC3(5) fluorescent dye, and fluorescence was monitored (emission, 670 nm; excitation, 544 nm) after addition of the nisin-subtilin hybrid peptide. (C and D) The SpaI-expressing strain B186.SB2 (C) and the NisI-expressing strain B168.SB1 (D) were analyzed as described for panel B. Stars indicate the time of addition of 2.5 μM DiSC3(5) fluorescent dye. As a control, 1 μM valinomycin (red) was added to the cells. The increase of relative fluorescence units (RFU) was monitored after addition of a 30 nM or 100 nM concentration of the hybrid molecule nisin1–17-subtilin18–32 (green) at 150 s, indicated by a plus sign.
As shown by the results in Fig. 3, the addition of 30 nM nisin to the SpaI-expressing strain resulted in immediate pore formation. In contrast, a concentration of 30 nM of the nisin1–17-subtilin18–32 hybrid peptide did not lead to any increase in fluorescence (Fig. 5C). If the same hybrid peptide was added to the NisI-expressing strain (B168.SB1) (Fig. 5D), already a concentration of 30 nM of the hybrid peptide led to a clearly measurable increase in fluorescence comparable to that without the immunity protein (Fig. 5B). The DiSC3(5) diffusion assay of the nisin1–17-subtilin18–32 hybrid peptide clearly showed that SpaI recognizes the C-terminal part of subtilin, thus mediating specific immunity against the nisin1–17-subtilin18–32 hybrid peptide. With respect to the antibiotic activity against the indicator strain Kocuria rhizophila ATCC 9341, the hybrid molecule was nearly (∼6 to 12 nM) as effective as wild-type entianin (subtilin) and nisin (∼3 to 6 nM). To further test whether the C-terminal lanthionine rings are important for SpaI immunity, mutants that lack either lanthionine ring D or E were analyzed (62). Surprisingly, SpaI also mediated immunity against these constructs comparable to that observed with entianin and the nisin1–17-subtilin18–32 hybrid peptide (Fig. S3). This shows that neither ring D nor ring E alone is essential for SpaI recognition. Notably, the antibiotic activities of ring D and E mutants against K. rhizophila were comparable to that of entianin (62). Unfortunately, mutants lacking both rings (D and E) could not be obtained (62).
Interaction of SpaI with the B. subtilis immunity transporter SpaFEG.
To further elucidate the role of SpaI and SpaFEG for immunity against entianin, both SpaI and SpaFEG were expressed in the subtilin-nonproducing strain B168 either alone or in combination with each other. The respective strains were subsequently tested for their immunity against entianin and nisin (Fig. 6 and Fig. S4).
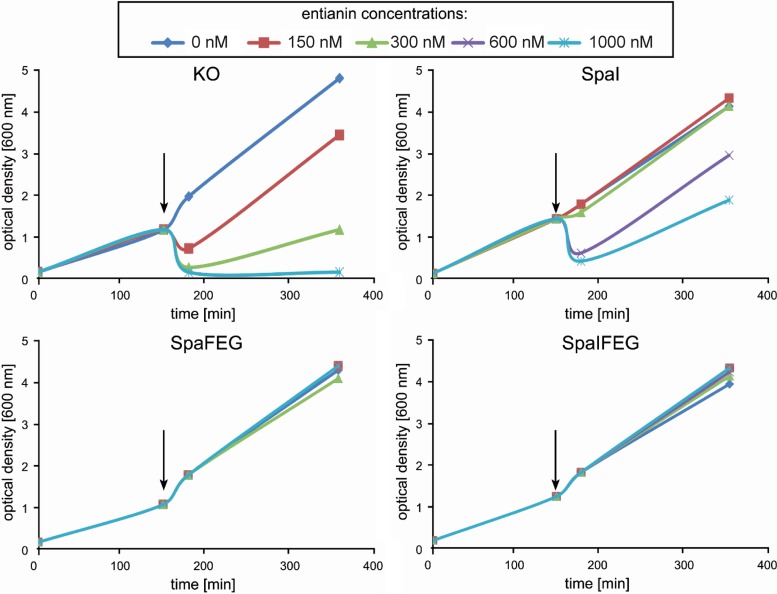
FIG 6.
Immunity mediated by SpaI, SpaFEG, and both expressed together. Growth curves of strains B168.KO (KO, knockout: without immunity proteins), B168.SB2 (SpaI), B2470.TM1 (SpaFEG), and B2470.TM2 (SpaIFEG) after addition of different concentrations of entianin. Arrows indicate the time point of entianin addition. Experiments were performed in triplicate.
The growth curves of the SpaI-expressing strain (B168.SB2) and the control strain without immunity (B168.KO) showed a dramatic difference after entianin addition. While the SpaI-expressing strain tolerated lantibiotic concentrations of 300 nM, the control strain already showed drastic growth inhibition at concentrations of 150 nM. This observation confirmed that SpaI was able to protect the cells from externally added entianin. However, the protection efficiency of SpaFEG (B2470.TM1) was much higher, conferring a tolerance of up to 1,000 nM, which is about three times higher than that of the SpaI-expressing strain. If SpaI was expressed in addition to SpaFEG, no additional effect of SpaI could be detected due to the strong immunity provided by SpaFEG alone.
As shown for nisin, two modes of action provide cellular toxicity. First, nisin binds to cell wall precursor lipid II, and second, nisin in complex with lipid II forms pores in the cellular membrane (53, 72). To test any impact of SpaI on membrane integrity, we analyzed the respective strains using DiSC3(5) diffusion assays (Fig. 7). Surprisingly, the tolerance against subtilin after SpaI expression was even stronger than that observed after SpaFEG expression. Furthermore, if compared to the results for SpaI expression, the simultanous expression of SpaI together with SpaFEG further increased the tolerance against entianin. This shows impressively that, with respect to membrane dissipation, SpaI has an important impact on immunity that is different from its minor efficiency in the growth tests. For SpaFEG, the application of 500 nM entianin showed a kinetics of DiSC3(5) release similar to that of the valinomycin control, whereas in the SpaIFEG strain, the DiSC3(5) release was delayed and 1,000 nM was needed to reach kinetics comparable to those of valinomycin.
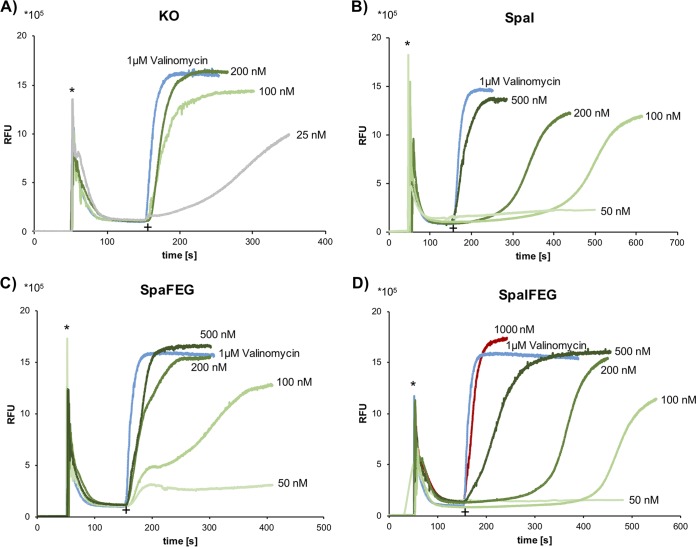
FIG 7.
Immunity mediated by SpaI, SpaFEG, and both expressed together. DiSC3(5) diffusion assay with the control strain B168.KO (without immunity proteins) (A) and strains expressing SpaI (B168.SB2) (B), SpaFEG (B2470.TM1) (C), and SpaIFEG (B2470.TM2) (D). Cells were incubated with DiSC3(5) fluorescent dye, and fluorescence was monitored (emission, 670 nm; excitation, 544 nm) after addition of membrane-active substances. Asterisks indicate addition of 2.5 μM DiSC3(5) fluorescent dye, and plus signs indicate the time when either 1 μM valinomycin (blue) or entianin at different concentrations (green shades, 50 to 500 nM; red, 1,000 nM; gray, 25 nM) was added.
Surprisingly, after the addition of 100 nM entianin to the SpaFEG strain, the DiSC3(5) release showed a two-step kinetics, starting with an immediate and drastic increase in fluorescence, which was attenuated before the DiSC3(5) release continued, reaching a maximal value comparable to that of the SpaI-expressing strain.
Whereas analysis of the growth curves did indicate a minor impact of SpaI in lantibiotic immunity, a significant role of SpaI became obvious with the DiSC3(5) diffusion assay.
SpaFEG was also tested for its ability to confer immunity against nisin (Fig. S4). Interestingly, in contrast to SpaI, SpaFEG also mediated increased tolerance against nisin. Unfortunately, an experiment performed vice versa with the NisFEG ABC transporter was not possible, as its expression in B. subtilis caused strongly reduced growth.
DISCUSSION
B. subtilis and L. lactis strains express two defense systems that protect the producing strains from the toxicity of their own synthesized lantibiotic. These are referred to as immunity proteins and comprise the lipoproteins SpaI (B. subtilis) and NisI (L. lactis), as well as the ABC transporters SpaFEG and NisFEG. Whereas it has been shown for both ABC transporters that they transport the respective lantibiotic from the membrane to the extracellular space (73, 38), the physiological function of SpaI and NisI was poorly understood. As shown here, both lipoproteins provide immunity to a nonproducing B. subtilis strain in a specific manner (Fig. 2 and 3). SpaI provides specific immunity against subtilin and subtilin-like lantibiotics, whereas NisI provides immunity against nisin. As previously shown, the positively charged amino acids within the N-terminal part of SpaI are unstructured and bind to the membrane similarly to the binding of the basic domain of NisI (49). Unfortunately, thus far, no direct physical interaction between SpaI and subtilin could be shown by NMR spectroscopy, as no suitable NMR buffer was found that kept both components together in solution (49). To follow the interaction of SpaI with subtilin, we applied LILBID-mass spectrometry (MS) (74), an emerging biophysical tool to determine the masses of large proteins, as well as protein complexes. Using LILBID analysis, we could clearly show the physical binding of subtilin to immunity protein SpaI (Fig. 1). Interestingly, monomers and dimers of SpaI were detected, and for both protein conformations, association with a single subtilin molecule was shown. However, using increased laser intensities, the SpaI dimer-subtilin complex was not detectable anymore, suggesting the decomposition of this complex. The stronger stability of the monomeric SpaI-subtilin complex indicates the predominance of this complex in vivo. Furthermore, this binding was more specific for subtilin. Although SpaI was able to bind nisin in minor but detectable amounts as well (Fig. 1 and Fig. S5), SpaI expression did not prevent nisin-mediated pore formation (Fig. 3). This shows that, under physiological conditions, SpaI specifically provides immunity against subtilin but not against nisin.
Characterization of the SpaI function is the last remaining puzzle in understanding lantibiotic biosynthesis and immunity. Despite the fact that SpaI and NisI provided increased immunity against their respective lantibiotics during growth analysis in vivo, the immunity provided by the SpaFEG transporter was much more pronounced (Fig. 6 and 7 and Fig. S4).
As shown for nisin, cellular toxicity is based on its binding to cell wall precursor lipid II and, additionally, by its forming pores in the cellular membrane in complex with lipid II (52, 53, 75, 58, 76, 56). While the growth tests were only suitable to follow the long-term immunity effects, the DiSC3(5) diffusion assays could directly follow the impact of immunity proteins on pore formation. Indeed, our current results show that delaying pore formation is the major physiological function of the lipoproteins SpaI and NisI. As expected, this also occurs in a specific manner, as pore formation provided by entianin was reduced upon SpaI expression, and vice versa, NisI was able to reduce nisin-mediated pore formation. As shown with a nisin-subtilin hybrid, the C-terminal part of subtilin/entianin is obviously important for its specific interaction with SpaI. Interestingly, neither lanthionine ring D nor ring E was important for the proper interaction with the lipoprotein. According to our findings, we assume that the positive charges within the C-terminal part of subtilin are attracted by the negatively charged membrane potential and, thus, promote its interaction with SpaI. This is consistent with our structural analysis of SpaI, assuming that, upon subtilin binding to lipid II via the first two lanthionine rings, the positive charges within the C-terminal part of subtilin are free to serve as binding site(s) for the highly negatively charged membrane proximal surface (49). Additionally, the fact that ramoplanin quenched subtilin-induced pore formation (Fig. 4) indicates that the immunity provided by SpaI is mainly due to its interaction with subtilin-lipid II complexes. This interaction prevents pore formation. Possibly, SpaI may even interact with existing pores, which would amplify the effect of a single SpaI molecule severalfold, comparable to the previously discussed mode of action (49). Similarly, a shielding mechanism where SpaI would close existing pores would also have an amplifying effect.
Remarkably, protection against lantibiotics epidermin and gallidermin depends on the transport provided by the ABC transporter and the LanH protein together with LanT (31, 32). Both lantibiotics, with only 22 amino acids, are much shorter than subtilin (32 amino acids) and nisin (34 amino acids). Due to their smaller size, epidermin and gallidermin do not form pores. Taking this together with our finding that the C-terminal amino acids of subtilin are needed to interact with SpaI (Fig. 5), we suggest that LanI expression is mainly important for pore-forming lantibiotics.
The impact of lipoproteins SpaI and NisI on cellular immunity remains a challenging question. On a first view, our growth analysis showed a considerable immunity effect of SpaI, but SpaFEG provided several-fold stronger immunity (Fig. 6). This suggested a major role of SpaFEG in providing cellular immunity. However, this picture changed completely when pore formation was examined (Fig. 7). Here, SpaI-mediated immunity was significantly more efficient than that provided by SpaFEG, which is quite the opposite of the findings with the long-term growth tests. Furthermore, combined expression of SpaI in a SpaFEG background increased entianin tolerance significantly more in the DiSC3(5) diffusion assay. This was in strong contrast to the results of the growth tests, where additional expression of SpaI did not strengthen SpaFEG-mediated immunity.
Surprisingly, the kinetics of DiSC3(5) release in a SpaFEG strain showed a two-step kinetics. This was most apparent when 100 nM entianin was applied (Fig. 7C). Obviously, in a first phase, the ABC transporter did not delay the DiSC3(5) response to entianin. However, after an adjustment period of approximately 30 s, the ABC transporter decelerated the DiSC3(5) release. This was completely different when SpaI was expressed together with SpaFEG, where a lag phase of more than 3 min was measured before DiSC3(5) was released. This delay was even longer than that provided by SpaI alone, revealing the physiological function of SpaI as a fast defense system that protects the cellular membrane immediately after its exposure to subtilin. Furthermore, the current results also show that, with respect to membrane protection, the immune response is much stronger if both immunity systems are expressed together.
Here, we analyzed the function of both lantibiotic immunity systems upon the provision of entianin and nisin externally. This is different from the situation during lantibiotic biosynthesis, which starts at the beginning of the stationary growth phase and gradually increases due to autoinduction. Possibly, upon increasing subtilin concentrations associated with the Gram-positive cell wall (38), the cellular membrane might use SpaI-mediated immunity as a first response until the SpaFEG transporter becomes active.
MATERIALS AND METHODS
Bacterial strains and plasmids used in this study.
Strains and plasmids used in this study are described in Table 1. Construction of the mutated spaS gene was performed as previously published (71), using pCG02 as a backbone. The respective plasmid containing the mutated subtilin structural gene was transformed into strains B15029.TSp01 and B15029 wild type as published previously (77). Plasmids expressing SpaI used for LILBID analysis were constructed as described in reference 78.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| B15029 | Wild type (Ent+); DSM 15029 | DSM |
| B6633 | Wild type (Sub+); ATCC 6633 | ATCC |
| TMB299 | [B168 amyE::pER605 (PyvcR-lacZ; –110–30) (Cmr)] | 27 |
| B168.KO | [B168 amyE::pER605 (PyvcR-lacZ; –110–30) (Cmr)] [thrC::pXT(Specr)] | This work |
| B168.SB1 | [B168 amyE::pER605 (PyvcR-lacZ; –110–30) (Cmr)] [thrC::pM1 Pxyl-nisI (–19–226) (Specr)] | This work |
| B168.SB2 | [B168 amyE::pER605 (PyvcR-lacZ; –110–30) (Cmr)] [thrC::pM2 Pxyl-spaI (–22–143) (Specr)] | This work |
| B2470 | [CU1065 liaI::pMutin (Eryr)] | 84 |
| B2470.SB2 | B2470 [CU1065 liaI::pMutin (Eryr), amyE::Pxyl-spaI (–22–143; Δ2–17) (Cmr) | 49 |
| B2470.SB3 | B2470 [CU1065 liaI::pMutin (Eryr), amyE::Pxyl-spaI(–22–143) (Cmr)] | 49 |
| B2470.TM1 | liaI:pMutin (Eryr) amyE::[xylR Pxyl-spaFEG Cmr] | This work |
| B2470.TM2 | liaI:pMutin (Eryr) amyE::[xylR Pxyl-spaIFEG Cmr] | This work |
| B15029.TSp32 | ΔetnS amyE::PspaS-spaS(W1I K2T E4I V12K Q17M) (Specr Neor) | This work |
| B15029.SK1 | ΔetnS amyE::PspaS-spaS(C26A) (Specr Neor) | 62 |
| B15029.SK3 | ΔetnS amyE::PspaS-spaS(C28A) (Specr Neor) | 62 |
| E. coli | ||
| DH5α | recA1 endA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44 φ80ΔlacZΔM15 Δ(lacZYA-argF)U169 | Life Technologies |
| M15 (pREP4) | Nals Strs Rifs Thi− Lac− Ara+ Gal+ Mtl− F− RecA+ Uvr+ Lon+ | Qiagen |
| K. rhizophila ATCC 9341 | Test strain for MIC determination | ATCC |
| S. cerevisiae CEN.PK2 | MATa/α ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 MAL2-8C/MAL2-8C SUC2/SUC2 | 85 |
| Plasmids | ||
| pH6TEVSpaI | MRSGH6-TEV SpaI (3–143) (Ampr) | 78 |
| pPK571 | MRSGH6-TEV SpaI (18–143) (Ampr) | 78 |
| pCG02 | bla amyE′ PspaS BamHI Neor ′amyE | 71 |
| pM1 | Pxyl-nisI[(−)19–226] in pXT via BamHI/EcoRI (Specr) | This work |
| pM2 | Pxyl-spaI(–22–143) in pXT via BamHI/EcoRI (Specr) | This work |
| pXT | pDG1731 derivative, integration vector for B. subtilis thrC locus, xylose-inducible promoter (Ampr Eryr Specr) | 86 |
| pTSp49 | bla amyE′ PspaS-spaS(W1I K2T E4I V12K Q17M) Neor ′amyE in pCG02 backbone | 71 |
| pSK1 | bla amyE′ PspaS-spaS(C26A) Neor ′amyE in pCG02 backbone | 62 |
| pSK3 | bla amyE′ PspaS-spaS(C28A) Neor ′amyE in pCG02 backbone | 62 |
| pXSpaFEG | amyE′ Pxyl-spaFEG (Ampr Cmr) ′amyE | This work |
| pXSpaIFEG | amyE′ Pxyl-spaIFEG (Ampr Cmr) ′amyE | This work |
Numerical ranges represent the expressed amino acid sequences of the constructs. Negative values are due to the upstream leader.
Production and purification of SpaI constructs for NMR analysis.
The coding sequences for the SpaI constructs (SpaI3–143 and SpaI18–143) were inserted into a modified pQE9 vector containing an N-terminal His6 tag followed by a tobacco etch virus (TEV) protease cleavage site. Following TEV protease cleavage, the expressed proteins contained two artificial residues at the N terminus, Gly and Ser for SpaI18–143 and Gly and Arg for SpaI3–143 (numbering refers to the wild-type SpaI sequence, starting at the lipidated cysteine with 1). The His-tagged SpaI was purified with a HisTrap HP column (GE Healthcare) in a first stage. After overnight TEV protease cleavage, the protease and the remaining uncleaved His-tagged protein were removed via an additional purification with a Ni-nitrilotriacetic acid (NTA) column. SpaI without a His tag was further purified using gel filtration.
LILBID-MS.
All samples were buffer exchanged into 20 mM Tris at pH 8.0 directly before the measurements with LILBID-MS. Buffer exchange and sample desalting were performed in Zeba Micro Spin desalting columns from Thermo Scientific, operating with a 7-kDa-cutoff filter. To equilibrate the desalting column, it was washed five times with the buffer at 1,500 rpm for 1 min, and the final sample buffer exchange was done for 2 min at 1,500 rpm. The sample concentration was around 15 µM. Amounts of 3 to 5 µl of buffer-exchanged samples were used for MS. To measure possible complex formation between SpaI and subtilin or nisin Z, the SpaI sample was desalted prior to the addition of the ligand in a 3-fold amount. One hour at 4°C was chosen to ensure possible complex formation, due to low binding kinetics. A piezo-driven droplet generator (MD-K-130; Microdrop Technologies GmbH, Norderstedt, Germany) was used to produce droplets of 30-µm diameter with a frequency of 10 Hz at 100 mbar. The droplets were transferred to high vacuum and irradiated by an infrared (IR) laser operating at 2.94 µm, a vibrational absorption wavelength of water. This led to an explosive expansion of the sample droplet and the release of ions, which were accelerated by a pulsed electric field and analyzed by a homebuilt reflectron time-of-flight (TOF) setup. LILBID settings have previously been published in detail (74).
This method generally offers some interesting advantages, as LILBID can be applied with a large variety of buffers and only 3 to 5 µl of the micromolar concentrated sample are needed. By using high concentrations of the analyzed ligand, the dilution factor of the protein is negligible. Furthermore, LILBID analysis takes only a few minutes, which overcomes sample precipitation during time-consuming NMR and isothermal titration calorimetry (ITC) analysis. Since there have been several successful LILBID measurements of small DNA-RNA-ligand complexes (79), as well as huge membrane-embedded molecules (80, 81), we also applied LILBID to detect the binding of immunity protein SpaI and its corresponding lantibiotic, subtilin. Data detection and processing were done using the software Massign (82). The MS spectra show averaged signals of 500 to 1,000 droplets. The spectra were normalized with the software OriginPro 2016. Figures were designed with Adobe Illustrator.
Purification and quantification of lantibiotics.
Purification of culture supernatants of lantibiotic producers and subsequent quantification of the proteins were performed as described previously (62).
Antibiotic activities of lantibiotic variants and hybrids using MICs.
The lowest concentration (MIC) that completely prevented growth of the cells was determined. Different indicator strains were used to determine the MICs, including B. subtilis 168, L. lactis MG1614 (83), and Kozuria rhizophila ATCC 9341. Indicator strains were cultivated under appropriate conditions. (B. subtilis and K. rhizophila were grown in LB medium [Formedium] at 37°C and 150 rpm. L. lactis was grown in M17 broth [Oxoid] supplemented with 0.5% glucose at 30°C without shaking). For the experiments, fresh overnight cultures of each strain were inoculated to a calculated optical density at 600 nm (OD600) of 0.001 in the respective medium. The indicator strain cultures were transferred in 2-ml portions into test tubes preloaded with the lantibiotic. Different drug concentrations were achieved by dilution of defined stock solutions. Growth was evaluated by visual examination after incubation at suitable temperature for 12 h.
DiSC3(5) diffusion assay.
Visualization assays of the transmembrane potential (Δψ) were performed after integration of the positively charged fluorogenic probe 3,3′-dipropylthiadicarbocyanine iodide [DiSC3(5)] into the membrane. B. subtilis cells were grown until the mid-exponential phase (OD600 of 0.9 to 1.5) and harvested at 16,000 × g for 5 min, and DiSC3(5) was added to the cells. Pore formation upon the addition of active antimicrobial peptides can be followed as soon as the leakage of the fluorogenic sample occurs. As a control, the dodecadepsipeptide valinomycin was used for all diffusion assays, as this ionophore is highly specific for potassium ions, finally resulting in the breakdown of the membrane potential. All DiSC3(5) diffusion assays were performed several times, and only one measurement is shown as a representative result, as each graph is composed of 3,000 to 4,000 measuring points and the associated error bars would overload the figure. For further details concerning the DiSC3(5) diffusion assay, see reference 62.
Determination of LanI-mediated immunity by growth analysis.
For the investigation of LanI-mediated immunity, SpaI- and NisI-expressing strains in the strain background of lantibiotic nonproducer B168 were used (B168.KO without immunity, NisI-expressing strain B168.SB1, and SpaI-expressing strain B168.SB2). The expression of LanI proteins was induced by the addition of 1% xylose to the cultivation medium (tryptone-yeast extract [TY]–0.3 M NaCl medium [0.8% {wt/vol} tryptone, 0.5% {wt/vol} yeast extract, 1.8% {wt/vol} NaCl]). A fresh overnight culture was used to inoculate 50 ml TY–0.3 M NaCl medium supplemented with 1% xylose to an OD600 of 0.1. Cultures were incubated at 37°C and shaken at 150 rpm until an OD600 of 0.9 to 1.1 was reached. At this time point, cultures were split into 2-ml samples and treated with the respective lantibiotic, with one sample left as an untreated control. After 30 min of incubation, the OD600 was determined and the values were normalized to those of a control without lantibiotic. For each strain, the control without added lantibiotic was set as 100%, whereas negative percental growth (indicating cell lysis) was not considered.
Determination of SpaIFEG-mediated immunity by growth analysis.
For further analysis of the immunity machinery, the ABC transporter SpaFEG (B2470.TM1) and the lipoprotein SpaI (B168.SB2) were expressed separately or in combination with each other (B2470.TM2) and tested against entianin and nisin. Expression of the immunity elements was induced by the addition of 1% xylose to the cultivation medium. Using a fresh overnight culture, a defined volume of TY medium supplemented with 0.3 M NaCl and 1% xylose was inoculated to an OD600 of 0.1 and incubated at 37°C (150 rpm). After reaching an OD600 of 0.9 to 1.1, cultures were split into 2-ml samples and treated with the respective lantibiotic (entianin or nisin). Subsequently, samples were taken at different time points for OD600 determination.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stefanie Düsterhus for excellent technical support and Carolin Hacker for critical reading of the manuscript and providing Fig. S2. We also acknowledge M. Karas for providing access to MS analysis.
This work was supported by grant number En 134/11-1 from the Deutsche Forschungsgemeinschaft (DFG) and the state of Hesse. Collaborator N.M. is supported by Cluster of Excellence Frankfurt (Macromolecular Complexes) and received funding from the European Research Council under the European Union’s Seventh Framework Programme (grant number FP7/2007-2013)/ERC grant agreement 337567. O.P. is supported by the DFG, collaborative research center 807, and J.M. is supported by grant number DFG-GRK 1986.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00534-19.
REFERENCES
- 1.Blaser M. 2011. Antibiotic overuse: stop the killing of beneficial bacteria. Nature 476:393–394. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 2.Willing BP, Russell SL, Finlay BB. 2011. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol 9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 3.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, et al. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnell N, Entian KD, Schneider U, Götz F, Zähner H, Kellner R, Jung G. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee S, Hansen JN. 1988. Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J Biol Chem 263:9508–9514. [PubMed] [Google Scholar]
- 6.Buchman GW, Banerjee S, Hansen JN. 1988. Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J Biol Chem 263:16260–16266. [PubMed] [Google Scholar]
- 7.Kaletta C, Entian KD. 1989. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J Bacteriol 171:1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnell N, Entian KD, Götz F, Hörner T, Kellner R, Jung G. 1989. Structural gene isolation and prepeptide sequence of gallidermin, a new lanthionine containing antibiotic. FEMS Microbiol Lett 49:263–267. doi: 10.1111/j.1574-6968.1989.tb03056.x. [DOI] [PubMed] [Google Scholar]
- 9.Kellner R, Jung G, Hörner T, Zähner H, Schnell N, Entian KD, Götz F. 1988. Gallidermin: a new lanthionine-containing polypeptide antibiotic. Eur J Biochem 177:53–59. doi: 10.1111/j.1432-1033.1988.tb14344.x. [DOI] [PubMed] [Google Scholar]
- 10.Kaletta C, Entian KD, Kellner R, Jung G, Reis M, Sahl HG. 1989. Pep5, a new lantibiotic: structural gene isolation and prepeptide sequence. Arch Microbiol 152:16–19. doi: 10.1007/BF00447005. [DOI] [PubMed] [Google Scholar]
- 11.Kaletta C, Entian KD, Jung G. 1991. Prepeptide sequence of cinnamycin (Ro 09-0198): the first structural gene of a duramycin-type lantibiotic. Eur J Biochem 199:411–415. doi: 10.1111/j.1432-1033.1991.tb16138.x. [DOI] [PubMed] [Google Scholar]
- 12.Dischinger J, Basi Chipalu S, Bierbaum G. 2014. Lantibiotics: promising candidates for future applications in health care. Int J Med Microbiol 304:51–62. doi: 10.1016/j.ijmm.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Düsterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJN, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kötter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nützmann H-W, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, et al. 2015. Minimum information about a biosynthetic gene cluster. Nat Chem Biol 11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen JN, Sandine WE. 1994. Nisin as a model food preservative. Crit Rev Food Sci Nutr 34:69–93. doi: 10.1080/10408399409527650. [DOI] [PubMed] [Google Scholar]
- 15.Younes M, Aggett P, Aguilar F, Crebelli R, Dusemund B, Filipič M, Frutos MJ, Galtier P, Gundert‐Remy U, Kuhnle GG, Lambré C, Leblanc J‐C, Lillegaard IT , Moldeus P, Mortensen A, Oskarsson A, Stankovic I, Waalkens‐Berendsen I, Woutersen RA, Wright M, Herman L, Tobback P, Pizzo F, Smeraldi C, Tard A, Papaioannou A, Gott D. 2017. Safety of nisin (E 234) as a food additive in the light of new toxicological data and the proposed extension of use. EFSA J 15:e05063. doi: 10.2903/j.efsa.2017.5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutowski-Eckel Z, Klein C, Siegers K, Böhm K, Hammelmann M, Entian KD. 1994. Growth phase-dependent regulation and membrane localization of SpaB, a protein involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol 60:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegers K, Heinzmann S, Entian KD. 1996. Biosynthesis of lantibiotic nisin. Posttranslational modification of its prepeptide occurs at a multimeric membrane-associated lanthionine synthetase complex. J Biol Chem 271:12294–12301. doi: 10.1074/jbc.271.21.12294. [DOI] [PubMed] [Google Scholar]
- 18.Kiesau P, Eikmanns U, Gutowski-Eckel Z, Weber S, Hammelmann M, Entian KD. 1997. Evidence for a multimeric subtilin synthetase complex. J Bacteriol 179:1475–1481. doi: 10.1128/jb.179.5.1475-1481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. 2015. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517:509–512. doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okeley NM, Paul M, Stasser JP, Blackburn N, van der Donk WA. 2003. SpaC and NisC, the cyclases involved in subtilin and nisin biosynthesis, are zinc proteins. Biochemistry 42:13613–13624. doi: 10.1021/bi0354942. [DOI] [PubMed] [Google Scholar]
- 21.Li B, Yu JP, Brunzelle JS, Moll GN, van der Donk WA, Nair SK. 2006. Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science 311:1464–1467. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- 22.Repka LM, Hetrick KJ, Chee SH, van der Donk WA. 2018. Characterization of leader peptide binding during catalysis by the nisin dehydratase NisB. J Am Chem Soc 140:4200–4203. doi: 10.1021/jacs.7b13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Meer JR, Polman J, Beerthuyzen MM, Siezen RJ, Kuipers OP, De Vos WM. 1993. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol 175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian KD. 1994. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol 60:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corvey C, Stein T, Düsterhus S, Karas M, Entian K-D. 2003. Activation of subtilin precursors by Bacillus subtilis extracellular serine proteases subtilisin (AprE), WprA, and Vpr. Biochem Biophys Res Commun 304:48–54. doi: 10.1016/S0006-291X(03)00529-1. [DOI] [PubMed] [Google Scholar]
- 26.Jordan S, Rietkötter E, Strauch MA, Kalamorz F, Butcher BG, Helmann JD, Mascher T. 2007. LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis. Microbiology 153(Pt 8):2530–2540. doi: 10.1099/mic.0.2007/006817-0. [DOI] [PubMed] [Google Scholar]
- 27.Rietkötter E, Hoyer D, Mascher T. 2008. Bacitracin sensing in Bacillus subtilis. Mol Microbiol 68:768–785. doi: 10.1111/j.1365-2958.2008.06194.x. [DOI] [PubMed] [Google Scholar]
- 28.Suntharalingam P, Senadheera MD, Mair RW, Lévesque CM, Cvitkovitch DG. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol 191:2973–2984. doi: 10.1128/JB.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein C, Entian KD. 1994. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl Environ Microbiol 60:2793–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegers K, Entian KD. 1995. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl Environ Microbiol 61:1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hille M, Kies S, Götz F, Peschel A. 2001. Dual role of GdmH in producer immunity and secretion of the Staphylococcal lantibiotics gallidermin and epidermin. Appl Environ Microbiol 67:1380–1383. doi: 10.1128/AEM.67.3.1380-1383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peschel A, Götz F. 1996. Analysis of the Staphylococcus epidermidis genes epiF, -E, and -G involved in epidermin immunity. J Bacteriol 178:531–536. doi: 10.1128/jb.178.2.531-536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aso Y, Okuda K-I, Nagao J-I, Kanemasa Y, Thi Bich Phuong N, Koga H, Shioya K, Sashihara T, Nakayama J, Sonomoto K. 2005. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci Biotechnol Biochem 69:1403–1410. doi: 10.1271/bbb.69.1403. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann A, Schneider T, Pag U, Sahl H-G. 2004. Localization and functional analysis of PepI, the immunity peptide of Pep5-producing Staphylococcus epidermidis strain 5. Appl Environ Microbiol 70:3263–3271. doi: 10.1128/AEM.70.6.3263-3271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein T, Borchert S, Kiesau P, Heinzmann S, Kloss S, Klein C, Helfrich M, Entian KD. 2002. Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol Microbiol 44:403–416. doi: 10.1046/j.1365-2958.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 36.Ra R, Beerthuyzen MM, de Vos WM, Saris PE, Kuipers OP. 1999. Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145:1227–1233. doi: 10.1099/13500872-145-5-1227. [DOI] [PubMed] [Google Scholar]
- 37.Stein T, Heinzmann S, Kiesau P, Himmel B, Entian K-D. 2003. The spa-box for transcriptional activation of subtilin biosynthesis and immunity in Bacillus subtilis. Mol Microbiol 47:1627–1636. doi: 10.1046/j.1365-2958.2003.03374.x. [DOI] [PubMed] [Google Scholar]
- 38.Stein T, Heinzmann S, Düsterhus S, Borchert S, Entian K-D. 2005. Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J Bacteriol 187:822–828. doi: 10.1128/JB.187.3.822-828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.AlKhatib Z, Lagedroste M, Fey I, Kleinschrodt D, Abts A, Smits SHJ. 2014. Lantibiotic immunity: inhibition of nisin mediated pore formation by NisI. PLoS One 9:e102246. doi: 10.1371/journal.pone.0102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.AlKhatib Z, Lagedroste M, Zaschke J, Wagner M, Abts A, Fey I, Kleinschrodt D, Smits SHJ. 2014. The C-terminus of nisin is important for the ABC transporter NisFEG to confer immunity in Lactococcus lactis. Microbiologyopen 3:752–763. doi: 10.1002/mbo3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takala TM, Saris PE. 2006. C terminus of NisI provides specificity to nisin. Microbiology 152:3543–3549. doi: 10.1099/mic.0.29083-0. [DOI] [PubMed] [Google Scholar]
- 42.Gebhard S. 2012. ABC transporters of antimicrobial peptides in Firmicutes bacteria—phylogeny, function and regulation. Mol Microbiol 86:1295–1317. doi: 10.1111/mmi.12078. [DOI] [PubMed] [Google Scholar]
- 43.Otto M, Peschel A, Götz F. 1998. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tü3298. FEMS Microbiol Lett 166:203–211. doi: 10.1111/j.1574-6968.1998.tb13891.x. [DOI] [PubMed] [Google Scholar]
- 44.Okuda K-I, Yanagihara S, Shioya K, Harada Y, Nagao J-I, Aso Y, Zendo T, Nakayama J, Sonomoto K. 2008. Binding specificity of the lantibiotic-binding immunity protein NukH. Appl Environ Microbiol 74:7613–7619. doi: 10.1128/AEM.00789-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okuda K-I, Yanagihara S, Sugayama T, Zendo T, Nakayama J, Sonomoto K. 2010. Functional significance of the E loop, a novel motif conserved in the lantibiotic immunity ATP-binding cassette transport systems. J Bacteriol 192:2801–2808. doi: 10.1128/JB.00003-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braun V, Wu HC. 1994. Lipoproteins, structure, function, biosynthesis and model for protein export, p 319–341. In Ghuysen J-M, Hakenbeck R (ed), Bacterial cell wall. New comprehensive biochemistry vol 27 Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 47.Kuipers OP, Beerthuyzen MM, Siezen RJ, de Vos WM. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem 216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 48.Halami PM, Stein T, Chandrashekar A, Entian K-D. 2010. Maturation and processing of SpaI, the lipoprotein involved in subtilin immunity in Bacillus subtilis ATCC 6633. Microbiol Res 165:183–189. doi: 10.1016/j.micres.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Christ NA, Bochmann S, Gottstein D, Duchardt-Ferner E, Hellmich UA, Düsterhus S, Kötter P, Güntert P, Entian K-D, Wöhnert J. 2012. The first structure of a lantibiotic immunity protein, SpaI from Bacillus subtilis, reveals a novel fold. J Biol Chem 287:35286–35298. doi: 10.1074/jbc.M112.401620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hacker C, Christ NA, Duchardt-Ferner E, Korn S, Göbl C, Berninger L, Düsterhus S, Hellmich UA, Madl T, Kötter P, Entian K-D, Wöhnert J. 2015. The solution structure of the lantibiotic immunity protein NisI and its interactions with Nisin. J Biol Chem 290:28869–28886. doi: 10.1074/jbc.M115.679969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuchs SW, Jaskolla TW, Bochmann S, Kötter P, Wichelhaus T, Karas M, Stein T, Entian KD. 2011. Entianin, a novel subtilin-like lantibiotic from Bacillus subtilis subsp. spizizenii DSM 15029T with high antimicrobial activity. Appl Environ Microbiol 77:1698–1707. doi: 10.1128/AEM.01962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruhr E, Sahl HG. 1985. Mode of action of the peptide antibiotic nisin and influence on the membrane potential of whole cells and on cytoplasmic and artificial membrane vesicles. Antimicrob Agents Chemother 27:841–845. doi: 10.1128/AAC.27.5.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sahl HG, Kordel M, Benz R. 1987. Voltage-dependent depolarization of bacterial membranes and artificial lipid bilayers by the peptide antibiotic nisin. Arch Microbiol 149:120–124. doi: 10.1007/BF00425076. [DOI] [PubMed] [Google Scholar]
- 54.Kordel M, Benz R, Sahl HG. 1988. Mode of action of the staphylococcinlike peptide Pep 5: voltage-dependent depolarization of bacterial and artificial membranes. J Bacteriol 170:84–88. doi: 10.1128/jb.170.1.84-88.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brötz H, Josten M, Wiedemann I, Schneider U, Götz F, Bierbaum G, Sahl HG. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol 30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 56.Medeiros-Silva J, Jekhmane S, Paioni AL, Gawarecka K, Baldus M, Swiezewska E, Breukink E, Weingarth M. 2018. High-resolution NMR studies of antibiotics in cellular membranes. Nat Commun 9:3963. doi: 10.1038/s41467-018-06314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breukink E, van Heusden HE, Vollmerhaus PJ, Swiezewska E, Brunner L, Walker S, Heck A JR, de Kruijff B. 2003. Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J Biol Chem 278:19898–19903. doi: 10.1074/jbc.M301463200. [DOI] [PubMed] [Google Scholar]
- 58.Hasper HE, de Kruijff B, Breukink E. 2004. Assembly and stability of nisin-lipid II pores. Biochemistry 43:11567–11575. doi: 10.1021/bi049476b. [DOI] [PubMed] [Google Scholar]
- 59.Wiedemann I, Benz R, Sahl H-G. 2004. Lipid II-mediated pore formation by the peptide antibiotic nisin: a black lipid membrane study. J Bacteriol 186:3259–3261. doi: 10.1128/JB.186.10.3259-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 61.Anderson RC, Hancock REW, Yu P-L. 2004. Antimicrobial activity and bacterial-membrane interaction of ovine-derived cathelicidins. Antimicrob Agents Chemother 48:673–676. doi: 10.1128/AAC.48.2.673-676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geiger C, Spieß T, Korn SM, Kötter P, Entian K-D. 2017. Specificity of subtilin-mediated activation of histidine kinase SpaK. Appl Environ Microbiol 83:e00781-17. doi: 10.1128/AEM.00781-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cavalleri B, Pagani H, Volpe G, Selva E, Parenti F. 1984. A-16686, a new antibiotic from Actinoplanes. I. Fermentation, isolation and preliminary physico-chemical characteristics. J Antibiot (Tokyo) 37:309–317. doi: 10.7164/antibiotics.37.309. [DOI] [PubMed] [Google Scholar]
- 64.Pallanza R, Berti M, Scotti R, Randisi E, Arioli V. 1984. A-16686, a new antibiotic from Actinoplanes. II. Biological properties. J Antibiot (Tokyo) 37:318–324. doi: 10.7164/antibiotics.37.318. [DOI] [PubMed] [Google Scholar]
- 65.Pallanza R, Scotti R, Beretta G, Cavalleri B, Arioli V. 1984. In vitro activity of A-16686, a potential antiplaque agent. Antimicrob Agents Chemother 26:462–465. doi: 10.1128/AAC.26.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Hare MD, Ghosh G, Felmingham D, Grüneberg RN. 1990. In-vitro studies with ramoplanin (MDL 62,198): a novel lipoglycopeptide antimicrobial. J Antimicrob Chemother 25:217–220. doi: 10.1093/jac/25.2.217. [DOI] [PubMed] [Google Scholar]
- 67.Cudic P, Behenna DC, Kranz JK, Kruger RG, Wand AJ, Veklich YI, Weisel JW, McCafferty DG. 2002. Functional analysis of the lipoglycodepsipeptide antibiotic ramoplanin. Chem Biol 9:897–906. doi: 10.1016/S1074-5521(02)00191-6. [DOI] [PubMed] [Google Scholar]
- 68.Brötz H, Bierbaum G, Leopold K, Reynolds PE, Sahl HG. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob Agents Chemother 42:154–160. doi: 10.1128/AAC.42.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stone KJ, Strominger JL. 1971. Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc Natl Acad Sci U S A 68:3223–3227. doi: 10.1073/pnas.68.12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Storm DR. 1974. Mechanism of bacitracin action: a specific lipid-peptide interaction. Ann N Y Acad Sci 235:387–398. doi: 10.1111/j.1749-6632.1974.tb43278.x. [DOI] [PubMed] [Google Scholar]
- 71.Spieß T, Korn SM, Kötter P, Entian K-D. 2015. Autoinduction specificity of lantibiotics subtilin and nisin. Appl Environ Microbiol 81:7914–7923. doi: 10.1128/AEM.02392-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bierbaum G, Sahl H-G. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol 10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 73.Stein T, Heinzmann S, Solovieva I, Entian KD. 2003. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J Biol Chem 278:89–94. doi: 10.1074/jbc.M207237200. [DOI] [PubMed] [Google Scholar]
- 74.Morgner N, Barth H-D, Brutschy B. 2006. A new way to detect noncovalently bonded complexes of biomolecules from liquid micro-droplets by laser mass spectrometry. Aust J Chem 59:109. doi: 10.1071/CH05285. [DOI] [Google Scholar]
- 75.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl H, de Kruijff B. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 76.Hsu S-T, Breukink E, Tischenko E, Lutters MA, de Kruijff B, Kaptein R, Bonvin A, van Nuland N. 2004. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol 11:963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 77.Spieß T, Korn SM, Kötter P, Entian KD. 2015. Activation of histidine kinase SpaK is mediated by the N-terminal portion of subtilin-like lantibiotics and is independent of lipid II. Appl Environ Microbiol 81:5335–5343. doi: 10.1128/AEM.01368-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Christ NA, Duchardt-Ferner E, Düsterhus S, Kötter P, Entian K-D, Wöhnert J. 2012. NMR resonance assignment of the autoimmunity protein SpaI from Bacillus subtilis ATCC 6633. Biomol NMR Assign 6:9–13. doi: 10.1007/s12104-011-9314-5. [DOI] [PubMed] [Google Scholar]
- 79.Morgner N, Hoffmann J, Barth H-D, Meier T, Brutschy B. 2008. LILBID-mass spectrometry applied to the mass analysis of RNA polymerase II and an F1Fo-ATP synthase. Int J Mass Spectrom 277:309–313. doi: 10.1016/j.ijms.2008.08.001. [DOI] [Google Scholar]
- 80.Morgner N, Kleinschroth T, Barth H-D, Ludwig B, Brutschy B. 2007. A novel approach to analyze membrane proteins by laser mass spectrometry: from protein subunits to the integral complex. J Am Soc Mass Spectrom 18:1429–1438. doi: 10.1016/j.jasms.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 81.Meier T, Morgner N, Matthies D, Pogoryelov D, Keis S, Cook GM, Dimroth P, Brutschy B. 2007. A tridecameric c ring of the adenosine triphosphate (ATP) synthase from the thermoalkaliphilic Bacillus sp. strain TA2.A1 facilitates ATP synthesis at low electrochemical proton potential. Mol Microbiol 65:1181–1192. doi: 10.1111/j.1365-2958.2007.05857.x. [DOI] [PubMed] [Google Scholar]
- 82.Morgner N, Robinson CV. 2012. Massign: an assignment strategy for maximizing information from the mass spectra of heterogeneous protein assemblies. Anal Chem 84:2939–2948. doi: 10.1021/ac300056a. [DOI] [PubMed] [Google Scholar]
- 83.Gasson MJ. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mascher T, Zimmer SL, Smith T-A, Helmann JD. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother 48:2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Entian K-D, Kötter P. 2007. 25 yeast genetic strain and plasmid collections, p. 629–666. In Stansfield I. (ed), Yeast gene analysis, 2nd ed, vol 36 Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 86.Derré I, Rapoport G, Msadek T. 2000. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37 degrees C. Mol Microbiol 38:335–347. doi: 10.1046/j.1365-2958.2000.02124.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.