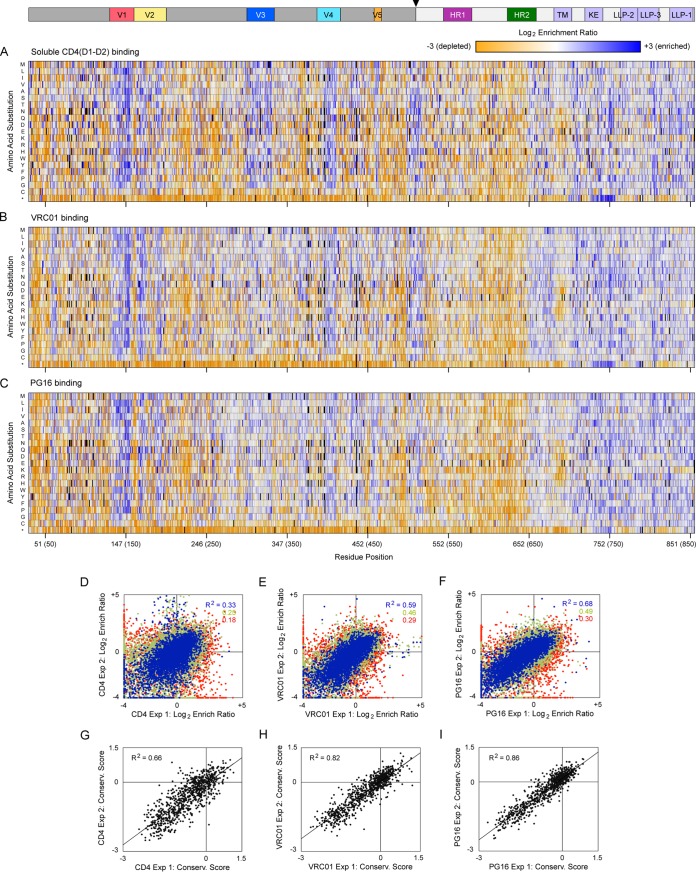
FIG 1.
Mutational landscapes of BaL Env interacting with protein ligands. (A to C) Merged data from in vitro evolution of three SSM libraries that together fully span the mature EnvBaL protein. The libraries were evolved by FACS for high binding signals to 200 nM CD4(D1-D2) (A), 5 nM VRC01 (B), and 2 nM PG16 (C). The Env sequence is on the horizontal axis (HXB2 numbering, BaL numbering in parentheses), and single amino acid substitutions are on the vertical axis. *, Stop codon. Log2 enrichment ratios are plotted from ≤–3 (depleted, orange) to 0 (neutral, white) to ≥+3 (enriched, blue). Mutations missing in the libraries (frequencies < 5 × 10−6) are black. The primary structure of gp120 (dark gray) and gp41 (light gray) is indicated above, with an arrowhead at the proteolysis site. Averages of two independent selection experiments are shown. (D to F) Correlation plots of mutation log2 enrichment ratios from independently replicated selections for high binding signals to soluble CD4 (D), VRC01 (E), and PG16 (F). Abundant mutations (frequencies > 2 × 10−4 in the naive library) are blue, mutations with moderate representation (frequencies between 5 × 10−5 and 2 × 10−4) are green, and rare mutations (frequencies between 5 × 10−6 and 5 × 10−5) are red. (G to I) Conservation scores were calculated by averaging the log2 enrichment ratios for all substitutions at each residue position. Conservation scores for libraries sorted for binding soluble CD4 (G), VRC01 (H), and PG16 (I) show agreement between replicate experiments.