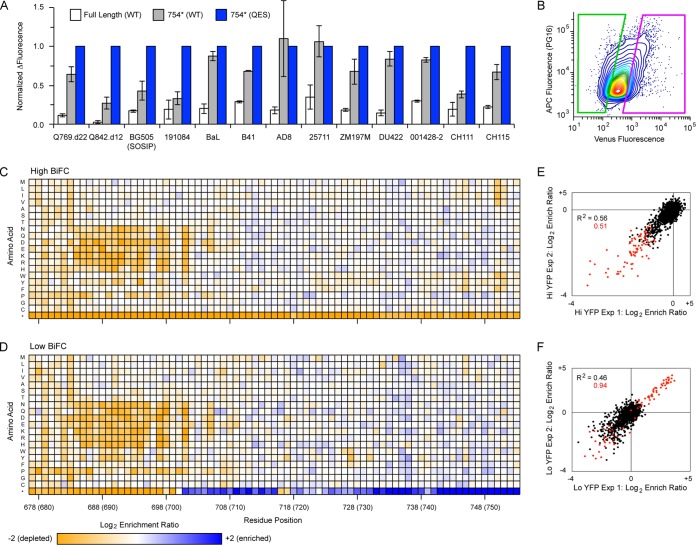
FIG 10.
Mutational landscape of Env TM and proximal cytosolic regions for associating with MA. (A) Wild-type Env (WT, gray) and QES variants of Env (blue) with premature stop codons at position 754 had enhanced expression compared to full-length WT Env (white). Transfected cells were stained with a high PG16 concentration (20 nM). The data are means (n = 2), with error bars showing the range. (B) Cells coexpressing 001428(753)-VC and MA-VN were gated for scatter properties, viability, low autofluorescence, and PG16 binding as described in Materials and Methods. The final collection gates are shown here. Cells were sorted for high BiFC (magenta, 15% of PG16-positive population) or low BiFC (green, 15% of PG16-positive population). (C) Mutational landscape of 001428 Env colocalizing with MA based on high BiFC. Log2 enrichment ratios are plotted from ≤–2 (depleted, orange) to ≥+2 (enriched, dark blue). Residue positions (HXB2 reference numbering, 001428 numbers in parentheses) are on the horizontal axis, and amino acid substitutions are on the vertical axis. Average of two independent replicates. (D) Mutational landscape of 001428 Env selected for low BiFC with MA. (E and F) Correlation plots for replicate experiments where cells expressing 001428 Env variants were sorted for high (E) or low (F) BiFC with MA. Nonsynonymous mutations are black; nonsense mutations are red.