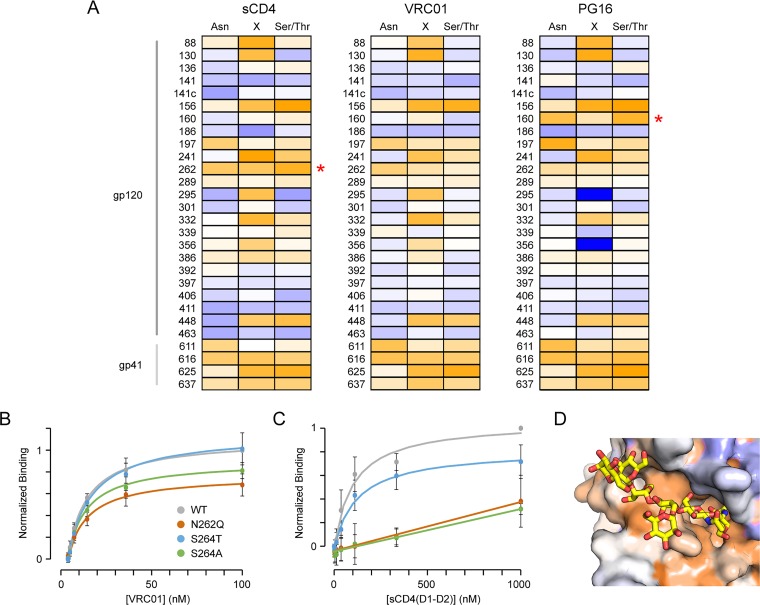
FIG 3.
Conservation of N-glycosylation sites. (A) Env libraries were evolved for binding soluble CD4, VRC01, and PG16 as indicated. Conservation scores of residues within N-glycosylation motifs are plotted from –2 (conserved, orange) to 0 (variable, white) to +2 (under selection for change, blue). Site numbering is based on Asn in the motif. Red asterisks indicate glycosylation sites preferentially conserved for binding only one of the three ligands. (B) Flow cytometry analysis of VRC01 binding to Expi293F cells expressing wild-type (WT, gray), N262Q (dark orange), S264T (blue), and S264A (green) Env variants (n = 4, means ± the standard deviations [SD]). (C) Binding of soluble CD4 to Env variants (n = 4, means ± the SD). (D) The N262 glycan (yellow sticks) occupies a cleft in the cryo-electron microscopy structure of CD4-bound Env (PDB 5VN3). The protein surface is colored by conservation score for CD4 binding, from ≤–2 (conserved, orange) to ≥+2 (under selection for change, dark blue).